Introduction
Small non-coding RNAs (sRNAs) are sequences involved in different constitutive as well as regulatory cell functions. Since the discovery of the first small RNA, lin-4 (Lee et al. 1993; Wightman et al. 1993), the list of small non-coding RNAs has increased, re-porting the participation of some of them in biological processes, such as the cutting and splicing of exons, genetic translation, development, differentiation, cell death, metabolic control, antiviral defense, and transcriptional and post-transcriptional regulation of gene expression (Gomes et al. 2013; van Wolfswinkel & Ketting, 2010; Winter et al. 2009). The function of many of these sRNAs is still unknown. Therefore, there is still an interest and a need for the detection, characterization and validation of their function.
The methodologies used to detect sRNAs face great challenges imposed by their size (≤ 200 nucleotides) and a low level of expression of the small RNAs, which in some cases does not exceed the detection threshold for the techniques used until now. For the aforementioned limitations, the most used methods for the detection of sRNAs (RT-qPCR and northern blot) need to be constantly optimized to improve sensitivity, specificity of detection, and quantification.
Northern blot is a direct methodology that permits the detection of the small RNAs, their precursory or intermediary sequences; the evaluation of their properties of expression; and establishment of their size (Wang & Yang 2010). Therefore, it continues to be widely used. The general northern blot process consists of total RNA extraction, separating it by size through electrophoresis, and transferring and immobilizing it on a solid surface (membrane). The probe with a complementary sequence to the RNA of interest is marked with radioactive and non-radioactive methods. Then, it is incorporated into the membrane that contains the immobilized RNA, and finally, the hybridization with the complementary sequence is detected.
Different protocols have been published for the northern blot methodology, which primarily vary in the design and marking of the probe. DNA probes marked with radioactive phosphorus (32P) are commonly used (Gurman S Pall & Hamilton, 2008; Varallyay et al. 2008). A new protocol for non-radioactive northern blot called LED (LNA-EDC-DIG) was reported by Kim et al. (2010), which integrates three separately developed methodologies that in turn modify three aspects of the process that determine the specificity and sensitivity of the northern blot. The modifications basically consist of replacing the DNA oligonucleotide probes with probes that contain locked nucleic acids (LNA). The immobilization of the RNA in the membrane, which is normally carried out with ultraviolet light (UV), is replaced with the use of EDC (1-Ethyl-3-(-3-dimethylaminopropyl) carbodiimide)). Additionally, the use of radioisotopes (32P), the most common method for marking probes, with which all of the risks of radioactivity must be assumed, is replaced by the use of digoxigenin (DG).
This new protocol increases the sensitivity and specificity in the detection of small RNAs by northern blot. Therefore, its use and implementation in studies of detection, characterization and functionality of new sR-NAs will be of great use. Due to this, the description of the implementation and modifications made by this protocol used in the detection of a small RNA, as well as the explanation and discussion of the theoretical foundations of the modifications posed, are the objective of this work.
Materials and Methods
Total RNA Extraction from Vero Cells
All RNA extractions were carried out with TRI-re-agent (Sigma™). The 4 x 106 Vero cells were quickly washed with 1X PBS to eliminate proteins and were resuspended in 1 ml of TRIzol. They were incubated for three minutes at room temperature (RT). Then, 200 mI chloroform (Merck™) were added and centrifugated at 5,000 revolutions per minute (rpm) for 15 minutes. The aqueous phase was taken and cold iso-propanol was added, and incubated for 10 minutes at -20 °C. Next, it was centrifugated again at 5,000 rpm for ten minutes using a microcentrifuge (Eppendorf™, 5810 R). The pellet was gently washed with 1 ml of 75% ethanol in a vortex mixer, centrifugated again at 10,700 rpm for five minutes, and then, the ethanol was left to dry at RT. Next, the pellet was resuspended in 100 |i of water treated with DEPC (Diethylpyrocar-bonate). The purity and concentration of the extracted RNA was determined with NanoDrop 2000 (Thermo Scientific™).
Non-radioactive Northern Blot
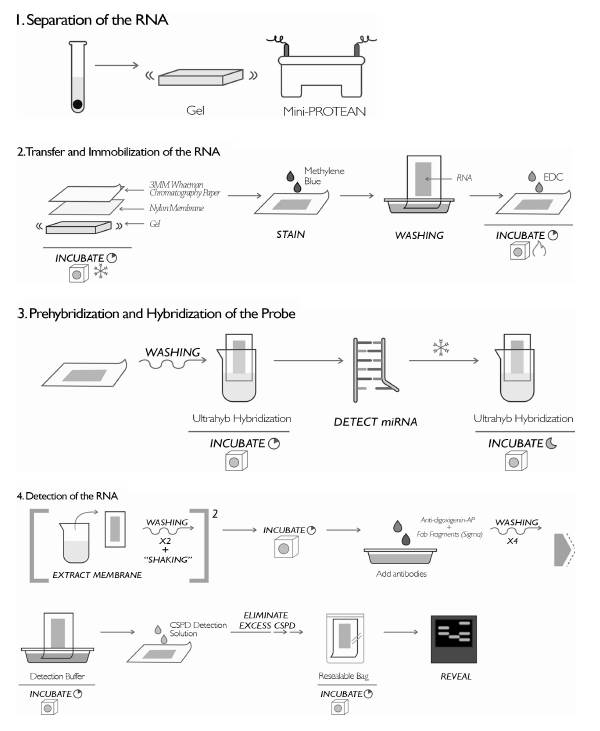
The extracted RNA was evaluated with non-radioactive northern blot. To do this, the protocol to detect mi-croRNAs (miRNAs), previously described by Kim et al. (2010), was modified, implemented and standardized. The protocol was developed in four steps (Figure 1): 1) Separation of the RNA in polyacrylamide gel; 2) Transfer and immobilization of the RNA; 3) Prehybridization and hybridization of the probe; and 4) Detection of the RNA. To implement the protocol, a specific and complementary probe to the small nuclear RNA (sn-RNA U6) was used, containing modified nucleotides (LNA) (Exiqon™). As a negative control, a probe also containing modified nucleotides (LNA) (Exiqon) was used, but in a "scramble" sequence.
Separation of the RNA: An electrophoretic run was made in 15% polyacrylamide gel (7 M urea danturant, Sigma™) using the Mini-PROTEAN system (Bio-Rad™, Hercules, CA, USA) and the molecular weight marker for miRNAs (New England BioLabs™ - microRNA Marker). Then 2 μg of the extracted RNA were mixed with loading buffer (Gel Loading Buffer II, Sigma™), the samples, and 5 μl of molecular weight marker. They were denatured at 95 °C for one minute, then loaded in gel and run applying 12.5 v/cm, in a Mini-PROTEAN (Bio-Rad™) electrophoresis chamber.
Transfer and immobilization of the RNA: Once the electrophoretic run was completed, the separate RNA was transferred to a positively charged nylon membrane (Roche™) using the Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad™) equipment. A sandwich-type assembly was made in this equipment with three sheets of 3MM Whatman chromatography paper (Whatman), the nylon membrane, the gel and finally three more sheets of chromatography paper. Then, 1X TBE (89 mM Tris base, 89 mM boric acid, and 2 mM EDTA were added. To ensure the transfer, this was carried out in a cold room at 15 V for 60 minutes. After the transfer time, it was evaluated whether there was transfer of the RNA to the membrane, staining it with methylene blue (0.02% methylene blue, 0.3 M NaOAc, pH 5.5) for three to ten minutes. The bands of RNA in the membrane were observed during this time, then the methylene blue was washed with distilled water. Subsequently, the RNA was immobilized in the membrane. To do this, a sheet of Whatman chromatography paper was moistened with EDC solution (24 ml EDC: 0.753 g 1-Ethyl-3-(-3-dimethylaminopropyl) carbodiimide -Sigma, 245 μl 1 Methylimidazole 12.5 M, and the final volume was made up by dilution with water treated with DEPC), and incubated for one hour at 65 °C.
Prehybridization and hybridization of the probe: After one hour of immobilization, the membrane was washed with deionized water. The Ultrahyb (Ambion) hybridization buffer solution was preheated to 65 °C, 15 ml of the buffer was added to each hybridization bottle, the membrane containing the immobilized RNA was added, and it was incubated for 30 minutes at 37 °C for prehybridization. The probes marked with digoxigenin and streptavidin to detect the miRNA and the molecular weight marker, respectively, were denatured at 95 °C for one minute. They were immediately put on ice and added to the hybridization bottle containing the membrane with the RNA, and incubated again at 37 °C throughout the night.
Detection of the RNA: To carry out the detection process, the membrane was withdrawn from the hybridization bottle, washed twice with low astringency buffer (2 x SSC with 0.1% (p/v) SDS) at 37 °C with agitation for 15 minutes; twice with high as-tringency buffer (0.1 x SSC with 0.1% (p/v) SDS) at 37 °C for five minutes; and once with wash buffer (1 x SSC) for ten minutes. Then, it was incubated at 37 °C for three hours in the block buffer (DIG wash and Block buffer set, Roche). Subsequently, the antibodies, anti-digoxigenin-AP, Fab fragments (Sigma) diluted at 1 : 15,000 and streptavidin (Pro-mega) at 1 : 4000 were added, and they were incubated for 30 minutes at RT. After incubating the antibodies, the membrane was washed with wash buffer (DIG wash and Block buffer set, Roche) four times for 15 minutes at RT. The membrane was incubated for five minutes in detection buffer (DIG wash and Block buffer set, Roche) at RT. Then, to the side of membrane that contains the RNA, the detection solution CSPD (Roche) was added in a dilution of 1 : 100. It was left at RT for five minutes. Then, the excess CSPD was eliminated from the membrane, put into a sealable heat resistant bag, and incubated for 15 minutes at 37 °C to proceed to its detection with a developing cassette and high sensitivity films. The films were exposed for one hour and 30 minutes to be subsequently developed.
Results and Discussion
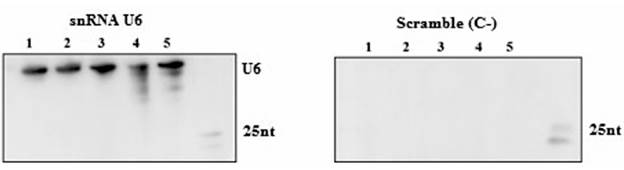
With the previously described protocol, a sign was observed when the RNA was evaluated with the specific probe of snRNA U6 (Figure 2); a small nuclear RNA involved in the assembly and functioning of the complex cell cutting and splicing, with an approximate size of 150 nucleotides (nt). When the same RNA was measured with the scramble negative control probe, no sign was observed (Figure 5). This indicates that the sign observed with the complementary probe to sn-RNA U6 is specific. The above demonstrates the sensitivity and specificity of the protocol in the detection of a constitutive cellular snRNA.
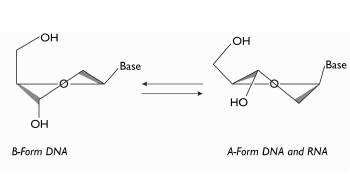
Figure 3 Outline showing the structures of the nucleic acids. B form in which the DNA is found. A form in which both DNA and RNA are found. Image taken and modified from Wengel (2009).
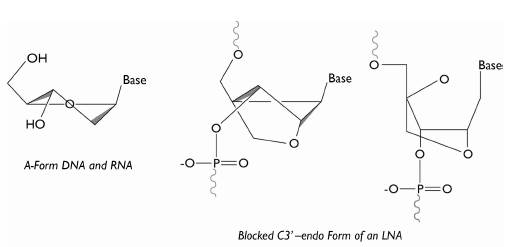
Figure 4 Illustration of the structure of an LNA. With the formation of the C3'-endo bond, the nucleotide remains in A form, the structural form in which the RNA is naturally found. Image taken and modified from Wengel (2009).
As initially mentioned, changes are proposed in this protocol in three aspects that contribute to the specificity, sensitivity and security of this technique, which are the type of probe, its marking, and the process of immobilizing the RNA in the membrane. To understand the reason behind the modifications made and for the later implementation and modification of this protocol, the theoretical foundations of the changes made to it are explained and discussed below.
Probe Design
When similar molecules to the nucleic acids are designed and synthesized, such as the probes used in northern blotting, three general characteristics must be taken into account. Firstly, the designed molecules must be efficient in oligomerization. Secondly, they must have high affinity and selective recognition of the complementary nucleic acid. This property can be measured by the melting point (MP) values; high values having less affinity and low values having more affinity. Thirdly, they must have good solubility in water (Wengel 1999).
To obtain probes with efficient oligomerization and high affinity in hybridization with the complementary chains of the nucleic acids, these must be designed so that the structure of each one of these probes has a structural form that is as similar as possible to the one naturally found in the nucleic acids (Petersen et al. 2002). Nucleic acids naturally have two kinds of structure: A form and B form. These formations occur because of the changes in the structure of the nucleo-tides' sugars (Wenge 1999). Ribose (sugar of RNA) is normally found in A form, while DNA can be found in either of the two structures, depending on the environment (ionic force and relative humidity) (Wengel 1999) (Figure 3).
In turn, the locked nucleic acids (LNA) are modified nucleotides. Their structure is characterized by having a methylene bridge that connects the 2'-oxygen with the 4'-carbon of the ribose (Figure 4). This bridge results in a blocked C3'-endo form, hindering the nucleotide from adopting another structure different to the A form; the structure in which RNA is naturally found (Gurman S Pall & Hamilton, 2008; Wengel 2009).
This results in the LNA probes used in the protocol proposed by Kim et al. (2010) having high affinity and thermal stability, represented in low MP values and greater specificity.
Immobilization of the RNA in the Membrane
The immobilization of the RNA in the membrane induced by UV occurs through crossover between the fractions of uracil and the positively charged amino groups present in the membrane (Sambrook et al. 2001). The process has three disadvantages, which notably affect the detection of small RNAs. Degradation of the functional groups occurs by UV (amino groups of the membrane) required for base pairing. The RNA are fragmented, decreasing their length and obstructing the maintenance of complete sequences, thus limiting the hybridization of the probes. As a final disadvantage, the number of bases involved cannot be controlled in the reaction induced by UV. Therefore, excessive crossover may occur, reducing the availability of bases for hybridization with a complementary probe.
Despite the above, the crossover between the RNA and membrane induced by UV is quick, cheap and efficient for RNA with more than 70 nt (Gurman S Pall & Hamilton, 2008). However, for the detection of small RNAs by hybridization, the pairing of the sequence is ideal so that it remains fully available for hybridization with the complementary probe (Gurman et al. 2008; Gurman et al. 2007). The crossover through the 5' end (different modifications can be found in the 3' end, and that is why the 5' end is preferred for a crossover) of the small RNAs, and not through the nitrogenated bases as occurs when using UV, is an alternative. It is possible to carry out this kind of crossover using 1-Eth-yl-3-(-3-dimethylaminopropyl) carbodiimide (EDC); a compound that has previously been used to crossover the 5' end of synthetic oligonucleotides in different solid substrates ( Gurman Singh Pall et al. 2007). The use of EDC in the immobilization of RNA in the membrane to detect miRNA arises as an alternative that notably improves sensitivity in the detection of these small molecules.
Marking of Probes
The northern blot transfer analysis using radioisotopes is the currently used methodology for the detection of sRNAs. However, this technique is generally expensive and costly, and implies environmental and occupational risks. Additionally, there is a justified and serious requirement not to promote the use of radioisotopes in Colombia. As an alternative, the digoxigenin (DIG) marking system is based on detection in the chemiluminescence process. This generates several advantages compared to the radioactive marking techniques, such as high sensitivity, short times of exposure, longer useful life and greater safety (Ramkissoon et al. 2006).
Conclusion
The growing need and interest in identifying, detecting, characterizing and validating our new sRNAs has been accompanied by the optimization and improvement of the methodologies used for these purposes, leading to protocols such as the previously described one. Knowing and understanding the theoretical foundations through which protocols are optimized, such as the one described in this work, contributes to development, to the proposal of new optimizations and to the creation of new protocols and methodologies that can be considered in the development of this kind of research.











 text in
text in