INTRODUCTION
Bioethanol can replace part of the consumption of traditional fuels such as oil or coal. It is possible to mix gasoline with bioethanol to offer savings in CO2 emissions of at least 50% compared to fossil fuels (Hernández Rodríguez & Hernández Zárate, 2008). However, be-cause bioethanol is traditionally manufactured with lignocellulose from crops such as corn, the price of food tends to increase, which becomes a reason to focus efforts on the use of lignocellulose from lignocellulosic waste for its production.
The lignocellulosic waste is composed of cellulose, hemi-cellulose and lignin and around 200,000 million tons are produced annually (Medina-Morales, Lara-Fernández, Aguilar, & de la Garza-Toledo, 2011; Saini, Saini, & Tewari, 2015). Colombia has a high production of lignocellulose, which can be used to produce bioethanol. If waste from Colombian industries such as citrus juice and rice husks were used, 136,045,500 L and 12,200,000 L of bioethanol would be obtained annually (Sánchez Riaño et al., 201 0). Taking into account the price of the gallon of ethanol of U$ 3.07 Colombian pesos (COP) (August 2018), approximately U$ 120,240,897 would be obtained (Sánchez Riaño et al., 2010).
Lignocellulose is currently hydrolyzed by mechanical breakdown followed by acid, alkaline and / or enzymatic hydrolysis (Persson, Tjerneld, & Hahn-Hágerdal, 1991; Cuervo, Folch, & Quiroz, 2009; Castillo et al., 2012). Of these three methods, the most promising by specificity and high recovery of glucose is enzymatic hydrolysis, with the disadvantage that the production of enzymes has high costs (Gao et al., 2008, G. Liu, Qin, Li, & Qu, 2013), which can be reduced if genetically modified microorganisms are used, to produce lignocellulolytic enzymes more efficiently in large quantities and in shorter times (Gao et al., 2008; Zelena, Eisele, & Berger, 2014).
Degradation of lignocellulose is carried out for a cellulolytic system composed of endoglucanase, exoglucanase and celubiosidase (Munjal, Jawed, Wajid, & Yazdani, 201 5).
According to the CAZy database, enzymes that degrade cellulose and other polysaccharides from plant cell wall, are classified into the glycose hydrolase families. In these GH families enzymes with the same enzymatic activity are not necessarily gathered, this classification is based on the sequence and structure of the protein, therefore families will understand different enzymatic activities (Busk, Lange, Pilgaard, & Lange, 2014).
The objective of this work was to evaluate the expression of endoglucanase fungal gene in E. coli BL21 (DE3) bacterium for the saccharification of lignocellulose residues, through the modification and cloning of eukaryotic genes of lignocellulolytic enzymes.
MATERIALS AND METHODS
Selection of eukaryotic genes encoding lignocellulolytic enzymes
The identifiers of the GH family were downloaded from the CAZy database containing the endoglucanase enzymes. Using these identifiers, the sequences of all the GH enzymes were downloaded from the GenBank, PDB and Uniprot databases (Berman et al., 2000, Magrane & Consortium, 2011).
Subsequently, a search was made of similar sequences to GH families enzymes using the BLAST program (Altschul et al., 1990), identifying functional protein do-mains annotated in Pfam (http://pfam.xfam.org/) (Finn et al., 2016) and Prosite (http://prosite.expasy.org/) (Sigrist et al., 2013). Later, domains such alignments were downloaded from Uniprot and each domain function were consulted and downloaded. Redundant sequences at 100% were eliminated with the CD-Hit software 4.6 (W. Li & Godzik, 2006).
Eukaryotic endoglucanase genes from fungi were select-ed. Selection criteria included presence of the specific domains for each group of enzymes, curation degree, presence of complete sequences in the databases, the size of the enzymes equal or greater than 60 kDa, ability to act extracellularly and the hydrolysis effectiveness of enzymes on specific substrates. The genes of the selected enzymes were annotated using InterproScan (Jones et al., 2014) to check their molecular function and bio-logical process.
Modification of selected genes
Selected genes CDS were obtained using the FGENESH software (Salamov & Solovyev, 2000). A codon preference adjustment was made for the standard organism Escherichia coli W311 0, genetically similar to E. coli BL21 (DE3) (Daegelen et al., 2009), using the Visual Gene Developer software (Gvritishvili et al., 2010; Jung & McDonald, 2011). Each of the modified sequences were checked using BLAST to verify that there was no loss of information and to corroborate that there was no loss of molecular function or biological process. Each sequence was annotated using Interproscan (Jones et al., 2014). Each of the genes was synthesized by GENEWIZ USA, in plasmid pUC57 for preservation, propagation and subsequent cloning in the expression vector pET151 TOPO.
3D modeling of modified proteins
3D modeling of the recombinant modified proteins was carried out with the I-Tasser software (J Yang et al., 2015), using as template the amino acid sequences of eglB modified genes in order to check their protein structure, its affinity with specific ligands and its molecular function. To perform 3D modeling quality were used Ramarchandran plot generated by the Swiss-PdbViewer software (Guex & Peitsch, 1997).
Molecular docking of 3d modeling versus pure sub-strates
Molecular docking was performed using SwissDock program (Schwede et al., 2003; Grosdidier et al., 2011), selecting the best 3D model with the C-score closest to 1. In the same way, the ligands were selected with the C -score closest to 1: (i) The ligand selected for docking with the 3D model of the endoglucanase enzyme was the cellobiose (CBI) in the PDB database (https://www3.rcsb.org/ligand/CBI).
The UCSF CHIMERA software (Pettersen et al., 2004) was used to elaborate the molecular analyzes and the docking graphs. OpenBabel software (Guha et al., 2006) were run to prepare the selected ligands according to the 3D topology to meet requirements of the SwissDock server.
Transformation of modified genes in E. coli Top10 cells for their conservation and propagation
The cloned and synthesized eglB gene in the plasmid pUC57 were transformed into E. coli Top10 competent cells for their stabilization and propagation, following manufacturer transformation protocol (FE Young & Spizizen, 1961; Hanahan, 1983; Inoue, Nojima, & Okayama, 1990, BD Hanahan et al., 1991). Then, positive transformants were selected and conserved by cryopreservation at -80 °C in 15% glycerol.
Positive transformants were inoculated in LB broth with ampicillin (100 lg / ml), to grow for 8 hours at 37 °C. Plasmid DNA extraction was performed following the standard procedure of alkaline lysis (Birnboim & Doly, 1979; Engebrecht, 1990). The results of the extraction were visualized in 1% agarose gel. Subsequently, con-centration and quality of extracted DNA was made using a nanodrop spectrophotometer (NanoDrop ND-2000 from Thermo Scientific).
Directional cloning of modified genes in expression vector pET151 TOPO
A standard PCR was run, preparing a reaction of 25 1L for each pUC57 plasmid DNA (Polymerase Platinum taq; DNA constructs of 1-10 ng per reaction; Primers 0.5 mM; dNTPs 0.2 mM, MgCl2 25 mM and 10X PCR Buffer). For the sequence of the eglB gene, the forward initiator Eg_CS_F and the reverse primer Eg_CS_R were used. Primers were designed according to the indications of the Champion™ pET151 Directional TOPO® Expression kit from Invitrogen (Invitrogen ™, 2010).
The thermal profile in the thermal cycler was constructed as described below: initial denaturation at 95° C for 5 minutes, followed by 35 cycles, each cycle with the cycle denaturation at 92° C for 2 minutes, then primer alignment to 57° C for 20 seconds and the extension at 72° C for 2 minutes a final extension of 72° C for 10 minutes was performed. PCR fragments was cloned following manufacturer cloning protocol in the TOPO vector (Invitrogen ™, 2010).
Constructs transformation in E. coli BL21 (DE3) for enzyme expression
The constructs were transformed into E. coli BL21 (DE3) cells (BD Hanahan et al., 1991; Invitrogen, 2013). Presence of genes were checked in colonies selected as positive transformants using standard colony PCR (Invitrogen ™, 2010) with appropriate primers.
Plasmid DNA extractions were performed for gene constructs in the pET151 TOPO vector, transformed in E. coli BL21 (DE3), and sequenced with the T7 promoter and T7 terminator primers. The integrity of the genes was corroborated using BLAST2seq and its orientation within the expression plasmid with the CLC program Main Workbench (www.qiagenbioinformatics.com/products/clc-main-workbench/).
Expression of the endoglucanase recombinant enzyme
Twenty microliters of cells transformed with eglB gene in E. coli BL21 (DE3) were inoculated on surface of CMC agar (10 g of CMC, 1 g of KH2PO4, 0.5 g of MgSO4.7H2O, 0.01 g of FeSO4.7H2O, 0.01 g of MnSO4.H2O, 0.3 g of NH4NO3 and 12 g of bacteriological agar, in one liter of distilled water with pH 7.0) (Ariffin et al., 2006).
Table 1 Selected CAZy families containing enzymes Endoglucanases.
| CAZy Family | Mechanlam | Proslte ID | CAZy NOTE | Members Number |
|---|---|---|---|---|
| CH5 (56 subfamilies) | Retaining | PS0O659 | Once known as cellulate family A; many members nave been assigned to subfamilies as described by Aspeborg et al. (2012) BMC Evol BioL 12(1):136 (PMID: 22992189). | 12233 Total 93 Archaea 9874 Bacteria 2139 Eukaryote 127 others |
| CH6 | Inverting | PS0O655 | formerly known as cellulase family B. The cellobiohydrolases of this famiry are widely believed to act processh ely from the non-reducing ends of cellulose chains to genérate ce oBiose | 1334 Total 1144 Bacteria 187 Eukaryota 16 others |
| CH7 | Retaining | NA | formerly known as cellulase family C. The cellobiohydrolases of this famiry act processivery from the reducing ends of cellulose chains to genérate cellobiose. This is markedly different from the IUBMB definition of cellobiohydrolases (EC 3.2.1.91), which act from the non-reducing ends of cellulose. | 5134 Total 10 Bacteria 5120 Eukaryota 4 others |
| CHS | Inverting | PS00812 | formerly known as cellulase family D | 3638 Total Bacteria 3809 11 Eukaryota 18 others |
| CH9 | Inverting | PS0O592 | formerly known as cellulase family E | 2771 Total 7 Archaea 1598 Bacteria 954 Eukaryota 212 others |
| GH12 | Retaining | NA | formerly known as cellulase family H | 552 Tota 90 Archaea 627 Bacteria 164 Eukaryota 1 Others |
| CH44 | Retaining | NA | formerly known as cellulase family J. Note that the stereochemistry of the reaction has recently been corrected: Kitago etal,/. Mol. Chem. 282(2007)35703-11 (PMID: 179057391 | 154 Total 149 Bacteria 3 Eukaryota 2 Others |
| GH45 | Inverting | PS01140 | formerly known as cellulase family K: distantty related to plant expansins | 385 Total 23 Bacteria 354 Eukaryota S Others |
| CH48 | Inverting | NA | formerly known as cellulase family L Some cellobiohydrolases of this famiry has e been reported to act from the reducing ends of cellulose (EC 3.2.1.-), while others have been reponed to opérate from the non-reducing ends to libérate cellobiose or cellotriose or cellotetraose (EC 3.2.1.-). This famiry also contains endo-processK e cellulases : EC 3.2.1.-), whose acthity is hard to distinguish from that of cellobiohvdrolases. | 1032 Total 995 Bacteria 34 Eukaryota 3 others |
| CH51 | Retaining | NA | NA | 2712 Total 20 Archaea 2486 Bacteria 165 Eukaryota 41 other: |
Then plates were incubated at 37 ° C for 48-72 hours. Then, agar was stained with 1% congo red and 1M NaCl. The hydrolysis rings were quantified using a calibrator. Measurement was taken from the outer end of the bacterial colony to the end of the hydrolysis halo on CMC agar (Meddeb-Mouelhi, Moisan, & Beauregard, 2014; Montoya et al., 2014).
Statistical analysis
To check which of the evaluated treatments had the highest degrading activity, an ANOVA analysis was per-formed, and a comparison of means was made using the Duncan method. Three technical replicas were made for each measurement. For each of four treatments evaluated (i) Treatment 1: negative control, E. coli BL21 (DE3) without transformation. (ii) Treatment 2: experimental control, Bacillus xiamenensis. (iii) Treatment 3: positive control, E. coli BL21 (DE3) transformed with plasmid pET151 TOPO without insert. (iv) E. coli BL21 (DE3) transformed with plasmid pET151 TOPO with the modified eglB gene.
RESULTS AND DISCUSSION
Ten families sequences belonging to CAZY database were downloaded from corresponding database (Figure 1), the identification catalytic regions from endoglucanase enzymes from literature search was done, as a result, an eukaryotic gene was selected from Aspergillus nidulans (Table 2).
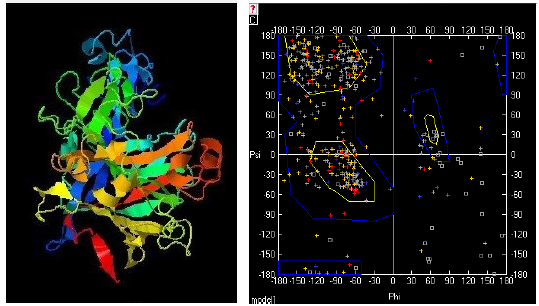
Figure 1 3D modeling for endoglucanase. (Left) l-TASSER best model for modified endoglucanase. C-score = 0.52 TM-score = 0.78 ± 0.09 RMSD = 5.9 ± 3.7 Á. (Right) Ramachandran graphic generated for the model.
Table 2 Description of the selected endoglucanase enzyme,
| Identifier | CAZy family | Enzyme description | Fungi | Interproscan GO Term Prediction |
|---|---|---|---|---|
| GenBank: AAM54071 Uniprot: Q5B7R2 | GH7 | Endo-beta-1,4-glucanasa: actividad extracelular. Su sustrato preferido es la Carboximetilcelulosa soluble. Tamaño 48 kDa. pH óptimo 5,5. Temperatura óptima 42 °C. (Lockíngton et al, 2002; Galagan et al., 2005; Bauer et al., 2006) | Aspergillus nidulans Asco mi cota | Función molecular: actividad hiclrolasa, hydroliza compuestos O-glicosilados |
Endoglucanase presented only one intron and the modification of the genes resulted in a sequence with different length from the original (Berg et al., 2002).
I-Tasser program predicted ten template structures, all of which are cellulase enzymes with a z-score greater than 1, which indicates that the alignments have a high quality (Roy et al., 2011). The program predicted 3 models for this enzyme, suggesting that the models used as a mold are remarkably similar to each other (Roy et al., 2011; J Yang & Zhang, 2016). The Ramachandran plot showed that the best model (Figure 1) presents most of the amino acids (which are not glycine or proline) in allowed areas, which indicates a good conformation (Mathews et al., 2002).
The models structurally similar to the problem protein (modified endoglucanase) were ten, which overlapped to the problem structure coincide almost in their entirety in their conformation, with TM-scores greater than 0.5, indicating the reliability of the prediction (J Yang & Zhang, 2016). The prediction of the function using the COACH server (Jianyi Yang et al., 2013) yielded 2 possible ligands: (i) Cellobiose. Identifier in the ChEBI, Pub-Chem and PDB databases: CBI (https://www3.rcsb.org/ligand/CBI). (ii) Beta-D-Glucose. Identifier in the ChEBI, PubChem and PDB databases: BGC (https://www3.rcsb.org/ligand/BGC).
The first of these substrates (ligands) corresponds to the cellulose monomer, which is the preferred substrate for endoglucanase enzymes and the second corresponds to the molecule that makes up the same cellobiose, so it also has affinity for this enzyme (Bohórquez Saval , 2012). This shows that the active site of the protein was correctly modeled (Roy et al., 2011). The residues in which the two similar ligands are located since both ligands share the amino acids located at positions 194, 218, 221, 232 and 376, which, as shown in Figure 2, are located in the central region of the protein.

Figure 2 Ligands identified for the modified endoglucanase protein. (Left) Docking site of modified endoglucanase. In blue, amino acids that share the three best ligands. In green, location of the CBI ligand. (Right) Binding sites of the best ligands selected by the l-Tasser for the endoglucanase 3D model.
The identifiers for the biological function or Enzyme Commission Numbers (E.C) found by the I-Tasser were two: (i) 3.2.1.4 which corresponds to the family of the cellulase enzymes or endoglucanases. (ii) 3.2.1.91, which is the identifier of cellulose 1,4-beta-celobiosidases (non-reducing end). Both identifiers correspond to enzymes that degrade cellulose. The consensus of the Gene Onthology prediction yielded as result for the molecular function the term with the number GO: 0016162 which corresponds to the cellulose activity 1,4-beta-celobiosidase and the GO term: 0008810, which identifies the cellulase activity. For the biological process, Gene Onthology gave the term GO: 0030245 that corresponds to the catabolic process of cellulose. For the cellular component, the term GO: 0005576 was obtained, which indicates that the protein functions in the extracellular region. The predictions of Gene Onthology yielded results that prove the biological process and molecular function of cellulose degradation. The above corroborates that the modified endoglucanase enzyme that was modeled by the I-Tasser can perform the degradation function of the cellulose polymer.
The molecular docking of the best model obtained from the I-Tasser against the specific ligands selected yielded mixed results, being able to compare the positions obtained in the SwissDock with the actual location of the ligands obtained by crystallography (G. Wu, Robertson, Brooks III , & Vieth, 2003) for endoglucanase enzyme, with a good result (Figure 3).
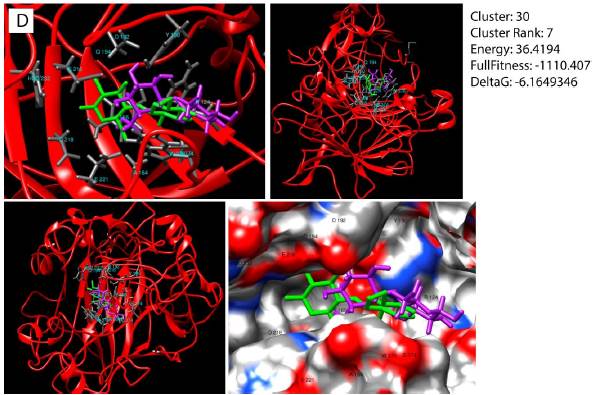
Figure 3 Best predicted docking for cellobiose ligand with the modified endoglucanase enzyme. Green colored molecule is the cellobiose ligand. Violet molecule is docking predicted by the SwissDock. In dark gray color the residues involved in the interaction with the ligand are appreciated.
Table 3 eglB gene sequence results obtained from nucleotide BLAST.
| Description | Max score | Total score | Query Coverage | E val. | Iclentity | Accession |
|---|---|---|---|---|---|---|
| Aspergillus nidulans endo-1,4-beta-glucanase (eg/Bl gene, complete cds | 1905 | 1905 | 93% | 0.0 | 98% | AF42002 |
| TPA Aspergillus nidulans FGSC A4 chromosome IV | 1853 | 1853 | 93% | 0.0 | 97% | BN00130 6.1 |
| Aspergillus nidulans FGSC A4 hypothetical protein AN3418.2 partial mRNA | 1853 | 1853 | 93% | 0.0 | 97% | XM.6559 30.1 |
| Synthetic construct 6HA'5-geloin gene, complete cds | 154 | 154 | 7% | 5e-33 | 100% | JX301693 |
PCR of the genes modified for cloning in pET151 TOPO vector, in the agarose gel, yielded bands of DNA fragments. eglB gene a band of ≈ 1300 bp was observed as expected. (Belancic et al., 1995, Lockington et al., 2002, Chávez et al., 2002, Ruhl et al., 2013).
From each of the colonies obtained in this transformation, the colony PCR was conducted, verifying that the colonies contained the respective amplicon with the expected sizes. The sequencing of PCR products resulted in the sequencing of the genes of interest for one eglB gene in pET151 in E. coli Top10 identified as E4T3.
The other samples sent for sequencing showed exceptionally low quality of the sequencing and were not analyzed. The only sample obtained for the eglB gene coincided with the sequence of the eglB gene from the organism Aspergillus nidulans by 99% (Table 2).
The analysis of sample E4T3, using the CLC Main Work-bench program, to check the orientation of the gene, showed that it was in the correct orientation within the expression vector pET151 TOPO.
In the three tests of enzymatic activity endoglucanase in E. coli BL21 (DE3), the eight replicas evaluated from the experimental control of Bacillus xiamenensis (positive control) presented halos of hydrolysis of the medium, between 2.45 and 4 mm, after development with Congo red (Meddeb-Mouelhi et al., 2014; Montoya et al., 2014), which was expected since their endoglucanase activity has already been proven (Lai et al., 2014). (Figure 4). The eight replicates of the E. coli BL21 (DE3) transformed with the eglB gene in pET151 TOPO presented hydrolysis halos of between 0.24 and 0.46 cm on the CMC agar in the presence of the IPTG expression inducer (Studier & Moffatt, 1986), similar to the hydrolysis rings obtained for experimental control.

Figure 4 hydrolysis haloes obtained from the third round of enzymatic activity of the endoglucanase recombinant enzyme. (a) Replies 1,2 and 3. Experimental control Bacilus xiamenensis. (b) Replicas 1,2 and 3. Negative control. E. coli BL21 (DE3) untransformed. (c) Replicas 1, 2 and 3. E coli BL21 (DE3) transformed with eglB gene in pET51 TOPO. (d) Replicas 1, 2 and 3. Positive control. E coli BL21 (DE3) transformed with pET151 TOPO vector.
On the other hand, the eight replicas of the negative control, of E. coli BL21 (DE3) without transformation, presented faint hydrolysis haloes that were almost imperceptible, showing little growth of colonies on the agar, which may indicate contamination with another bacterium that is able to grow in the agar taking advantage of the sugars of the CMC, since this is a semi-selective agar, whose only carbon source is the CMC (Meddeb-Mouelhi et al., 2014), which cannot be metabolized by the E.coli strain BL21 (DE3) untransformed. As regards the eight replicates of the positive control of E. coli BL21 (DE3) transformed with the plasmid pET151 TOPO with-out insert, they did not present any hydrolysis halo. (Figure 16). These results suggest that the endoglucanase enzyme was expressed correctly in E. coli BL21 (DE3) cells after the first transformation.
The statistical analysis was performed for the semiquantitative measurement of the hydrolysis of the recombinant endoglucanase enzyme. The ANOVA analysis resulted in statistical significative differences (p value <0.0001). Duncan's analysis showed the four treatments, grouped into three different groups. The values obtained by the comparison for each treatment were the following: (i) Treatment 1: 0 cm. (Group C) (ii) Treatment 2: 0.29625 cm. (Group B) (iii) Treatment 3: 0 cm. (Group C) (iv) Treatment 4: 0.39375 cm. (Group A). The treatment iv with the highest hydrolysis halo was E. coli BL21 (DE3) transformed with the modified eglB gene in the plasmid pET151 TOPO (Group A), followed by the experimental control Bacillus xiamenensis (Treatment 2, Group B), which presented the second largest hydrolysis halos.
CONCLUSIONS
The 3D structures predicted by the I-Tasser proved to be very close to the existing structures of similar enzymes in the PDB database, being able to demonstrate that they were structures that were correctly folded that did not undergo major changes in their conformation due to the elimination of introns and modification of codons, which shows that this process did not affect in silico the functionality of the enzymes. Regarding the ligation sites, predicted with the SwissDock, were highly conserved sites modeled correctly in the modified enzymes. The results of 3D modeling and docking allowed to corroborate in a preliminary way that these did not suffer important changes that affected their functionality during the process of gene modification.
The sequencing also allowed us to conclude that eglB genes cloned within the E. coli Top10 and E. coli BL21 (DE3) cells underwent very little alteration and remained almost identical to the originally modified genes. T7 promoter and the restriction analysis carried out for each of the constructs, allow to elucidate whether the genes were in the correct orientation within the plasmid for the eglB gene, which was in the correct orientation.
The expression of the modified endoglucanase enzyme was achieved in the expression cells E. coli BL21 (DE3), in soluble and functional form, demonstrated by hydrolysis of the CMC substrate. Cells from E. coli BL21 (DE3) transformed with the modified eglB gene in the pET151 TOPO vector were shown to form larger hydrolysis rings than the experimental control Bacillus xiamenensis. The expression and activity of enzymes sequences have to overcome many troubles such as possible incorrect orientation within the expression plasmid; the possible formation of inclusion bodies that could have prevented the secretion to the outside of the cell due to its size and recombinant nature; the natural defense mechanisms of E. coli to avoid toxic levels of enzymatic cofactors within its cytoplasm, which may prevent the correct folding of enzymes; the possibility that there has been an incorrect folding enzyme due to its recombinant nature. All these problems can be partially avoided if bioinformatics is applied to change amino acids without alter functionality and affinity for substrate. This is a step forward to produce endoglucanase in vitro, with a high efficiency because bacteria low cost and high producing system expression.














