1. INTRODUCTION
Escherichia coli is a gram-negative bacillus that belongs to the Enterobacteriaceae family and is considered to be a member of the extraintestinal pathogenic E. coli (ExPEC) group. Some strains are potentially pathogenic and can cause serious diseases in humans and animals, such as urinary tract infection, neonatal meningitis, septicemia, and in some cases can cause the death (Wang et al., 2002; Kaper et al., 2004). Avian pathogenic E. coli (APEC), belongs to the strains that produce an extraintestinal infection in chickens, turkeys, and other types of birds, called colibacillosis (Wang et al., 2002), characterized by an initial respiratory disease, followed by a systemic infection that induces fibrinous lesions in different organs, causing airsacculitis and associated pericarditis, perihepatitis, as well as, cellulitis, peritonitis, and fatal septicemia (De Carli et al., 2015; Guabiraba & Schouler, 2015). APEC is considered one of the main causes of economic losses due to morbidity, mortality that in some cases can reach almost 20%, and discard of poultry carcasses worldwide (Paixão et al., 2016; Schouler et al., 2020; Mageiros et al., 2021).
The E. coli strains responsible for these extraintestinal infections in birds have several genes that encode virulence factors related to processes such as adhesion, invasion, iroN acquisition systems, hemolysis, immune response evasion, resistance to antibiotics, and toxins production (Dziva & Stevens, 2008; Schouler et al., 2020). These virulence genes are encoded in the genome of E. coli and/or its plasmids (Dozois et al., 2003). Due to the fact that these genes are not present in all isolates, in some cases, one or multiple virulence genes can occur, some of these detected in isolates from healthy birds. Although it has been reported that the pathogenicity of APEC strains is likely to be determined by the presence of at least five virulence genes (Dziva & Stevens, 2008; De Carli et al., 2015; Subedi et al., 2021). The specific virulence APEC genes are less described than in human ExPEC pathotypes. In consequence, it has been difficult to understand whether it is just one or a combination of virulence factors associated with the strains that cause the disease (Mageiros et al., 2021).
In the present study, the evaluation of 13 virulence factors of APEC (iroN, hlyF, iss, iutA, frz, vat, sitA, KpsM, sitD, fimH, pstB, sopB, and uvrY) was performed. They were selected from to a review of the virulence genes that have been most frequently reported in APEC isolates in other countries (Dozois et al., 2003; Ewers et al., 2005; Johnson et al., 2008). The salmochelin siderophore receptor gene, iroN, facilitates the chelation of iroN in the host (Bäumler et al., 1998). A putative avian hemolysin gene, hlyF and an aerobactin-siderophore receptor gene, iutA, both contributing to iroN absorption (van der Westhuizen & Bragg, 2012). Besides, there is an episomic increase of the serum survival gene, iss, which helps with resistance to host serum (Derakhshandeh et al., 2009). The frz operon promotes bacterial fitness under stressful conditions (Rouquet et al., 2009). The vat gene encodes a vacuolating autotransporter toxin, that could be related to agglutination, biofilm formation as well as virulence (Parreira & Gyles, 2003). The sitA and sitD genes are part of the sitABCD system, classified as a bacterial iroN transporter (Runyen-Janecky et al., 2003).
The capsule formation transporter gene, KpsM, encodes a polysaccharide protein.
Transporter for the formation of protective capsule (Pavelka et al., 1991). The type 1 fimbrial adhesion gene, fimH, contributes to the protection from the host heterophile antibodies (Mellata et al., 2003). The pstB gene which is part of the pstSCAB operon, has been shown to increase resistance to polymyxin, rabbit serum, and acid shock (Lamarche et al., 2005). The sopB-encoded plasmid division protein is common in several plasmids, including pAPEC-1 (GenBank accession number CP000836), pAPEC-O1-ColBM (GenBank accession number DQ381420), and pVM01 (GenBank accession number EU330199), associated with virulence traits in APEC (Runyen-Janecky et al., 2003). And a transcriptional regulatory gene of iroN uptake genes in APEC, uvrY, which is involved in the regulation of carbon metabolism and contributes to its virulence (Li et al., 2008).
On the other hand, antibiotics have been for a long time the first line of defense to prevent APEC, but they have lost their clinical efficacy as bacterias have become increasingly resistant to treatment due to their irrational use, and at the moment there is no effective vaccine because of the multiple serotypes involved (Moriel et al., 2010; Tuntufye et al., 2012). In recent years, it has been reported a high prevalence of multidrug-resistant strains among causal isolates of colibacillosis (Hazam et al., 2019; Song et al., 2020), as well as, outbreak alerts where multi-resistant APEC has been identified (Solà-Ginés et al., 2015; Gao et al., 2018). It seems that these strains are not only facilitating the transmission and dissemination of drug resistance and other virulence factors between bothanimal and human pathogens, but also they could increase antimicrobial resistance in other organisms (pathogenic and nonpathogenic) within gastrointestinal tract of the chicken (Tello et al., 2012; Kabiswa et al., 2018). In addition, APEC does not only share identical serotypes with human pathogens but also specific virulence factors, therefore their zoonotic potential is under consideration and is highly concerning (Berg et al., 2017; Nadimpalli et al., 2019; Umair et al., 2019; Zhuge et al., 2019).
Genetic diversity is an obstacle for the identification of common properties, which could be used as a basis for diagnostic methods and vaccination, this challenging condition is related to the arduous control of avian colibacillosis. Due to this, the identification and characterization of virulence genes have emerged as a need to develop Specific therapeutic targets that could contribute to the implementation of new strategies for treatment.
In Colombia, to date, exists some reports related to antibiotic resistance which in this case has an intricate relation to virulence, however, no studies have been conducted evaluating the presence of virulence factors in isolated strains of APEC, which makes it difficult to determine specific strains for vaccine development, the effectiveness of antibiotics or even the prevalence among infections in the community. The present work aimed to characterize the genetic profile of some virulence factors of different isolates of avian E. coli in Caloto, Cauca, Colombia.
2. MATERIALS AND METHODS
2.1 Sample collection
A total of 47 E. coli strains were isolated from the cultures of oviduct, lung, liver, coelomic cavity and yolk sac samples taken from animals, diagnosed with bursitis in the region of Caloto, Cauca, Colombia.
2.2 Isolation and bacterial identification
The bacteria were isolated on MacConkey agar, subsequently, they were identified using biochemical tests such as TSI, use of citrate, SIM and LIA, and finally, 3 isolated colonies of each strain were placed in sterile PBS for DNA extraction. All isolates were initially identified through 16s ribosomal amplification.
2.3 DNA extraction
The DNA was extracted using Chelex® 100. For this method, 100 μl of the culture was centrifuged at 12,000 rpm for 5 minutes, then the supernatant was removed and the pellet resuspended in 100 μL of miliQ water, then vortexed for 30 seconds and added 100 μL of Chelex 10%. The samples were incubated at 99°C for 20 minutes in a Vortex at 70 rpm, left at room temperature for 10 minutes, and then centrifuged at 12,000 rpm for 10 minutes. Finally, the supernatant was transferred to a new 1.5 ml tube and stored at -20°C until its use.
The amount of DNA in each of the extractions was determined using a Nanodrop
(ND1000, Thermo Scientific, Wilmington, USA). For the evaluation of the integrity of the DNA, electrophoresis was performed using 1% agarose gel, containing 0.5 μg/ml of ethidium bromide. Images were acquired using photodocument of Chemidoc gels (Biorad) and the quantityOne software.
2.4 Oligonucleotide design
The genes selected to amplify in this study were chosen according to literature reports as previously mentioned. There were used primers for each of the virulence factors previously reported. The size of the amplified fragments was between 198 bp and 843 bp (Table 1).
Table 1 List of primers for each gene, sequence, size of each fragment to be amplified, concentration of the primer used for each multiplex PCR, and the reference from where the sequences were taken.
| Gene Multiplex 1 | Sequence 5´-3´ | Reference | Size Fragment | Concentración µM |
|---|---|---|---|---|
| iroN F | AATCCGGCAAAGAGACGAACCGCCT | (Johnson et al., 2008) | 553 pb | 0.25 |
| iroN R | GTTCGGGCAACCCCTGCTTTGACTTT | 0.25 | ||
| hlyF F | GGCCACAGTCGTTTAGGGTGCTTACC | (Johnson et al., 2008) | 450 pb | 0.25 |
| hlyF R | GGCGGTTTAGGCATTCCGATACTCAG | 0.25 | ||
| Iss F | CAGCAACCCGAACCACTTGATG | (Johnson et al., 2008) | 323 pb | 0.25 |
| Iss R | AGCATTGCCAGAGCGGCAGAA | 0.25 | ||
| iutA F | GGCTGGACATCATGGGAACTGG | (Johnson et al., 2008) | 302 pb | 0.25 |
| iutA R | CGTCGGGAACGGGTAGAATCG | 0.25 | ||
| Gene Multiplex 2 | ||||
| frz-FP | GAGTCCTGGCTTGCGCCGTT | (van der Westhuizen & Bragg, 2012) | 843 pb | 0.5 |
| frz-RP | CCGCTCCATCGCAGCCTGAA | 0.5 | ||
| vat-FP | CGCTTCAGGTGCGCTGACCA | (van der Westhuizen & Bragg, 2012) | 498 pb | 0.5 |
| vat-RP | AAGGGAGACGATGCCCGCCT | 0.5 | ||
| sitA-F | CGCAGGGGGCACAACTGAT | (Sabri et al., 2008) | 661 pb | 0.5 |
| sitA-R | CCCTGTACCAGCGTACTGG | 0.5 | ||
| KpsM-FP | CAGCCTCGCGGCTTAGCTCC | (van der Westhuizen & Bragg, 2012) | 335 pb | 0.5 |
| KpsM-RP | TGCACGCGCACTGCTTGAGA | 0.5 | ||
| Gene Multiplex 3 | ||||
| sitD-F | CTGTGCGCTGCTGTCGGTC | (Sabri et al., 2008) | 571 pb | 0.6 |
| sitD-R | GCGTTGTGTCAGGAGTAC | 0.6 | ||
| fimH-FP | GGATAAGCCGTGGCCGGTGG | (van der Westhuizen & Bragg, 2012) | 331 pb | 0.6 |
| fimH-RP | CTGCGGTTGTGCCGGAGAGG | 0.6 | ||
| pstB-FP | CGCGCTCGTCCATGTCAGCA | (van der Westhuizen & Bragg, 2012) | 198 pb | 0.6 |
| pstB-RP | CGGAACAGCGTGCGGAAGGT | 0.6 | ||
| sopB-FP | ACGACCCCCAGGGAACAGCA | (van der Westhuizen & Bragg, 2012) | 797 pb | 1 |
| sopB-RP | TGCAGCTGGTGCCGATGACG | 1 | ||
| uvrY-FP | TGAGTGCGATTCGTTCTGTC | (Herren et al., 2006) | 286 pb | 0.6 |
| uvrY-RP | TCTCCGCATTACACAGACCA | 0.6 |
The PCR reaction mixtures were subjected to the following conditions in a Biorad 1000 thermocycler (Biorad, CA, USA): denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 30 sec, 63°C for 30 sec and 72°C for 3 min and a final cycle of 10 min at 72°C.
The reaction mixture contained a final volume of 25 μl: 2 μl of DNA, 2.5 μl of the buffer, 0.25 mM of dNTP (1.2 μl), 3 mM MgCl2 (1.5 μl), oligonucleotides 0.25-1 μM (0.625 μl) Taq polymerase 0.2 μl and nuclease-free H2O 13,9 μl. The products were visualized by electrophoresis using 4% agarose gel (w/v) in a 1X TAE buffer run at 100 V for 80 minutes.
3. Analysis of results
The results were analyzed to generate the following virulence profiles according to the number of genes related to virulence factors found in the different strains: profile A, isolates that had between 10 and 13; profile B, those that had between 5 and 9 and in profile C, those who had 4 or less virulence factors.
4. RESULTS
A total of 47 E. coli strains were isolated from the cultures of oviduct, lung, and liver. Samples. A bacterial pool containing E. coli strains with different virulence factors was used as a positive control since no strain had all the virulence factors in study. The three multiplex PCRs were run with the DNA pool to ensure that under the proposed conditions the different genes were amplified (Fig. 1).
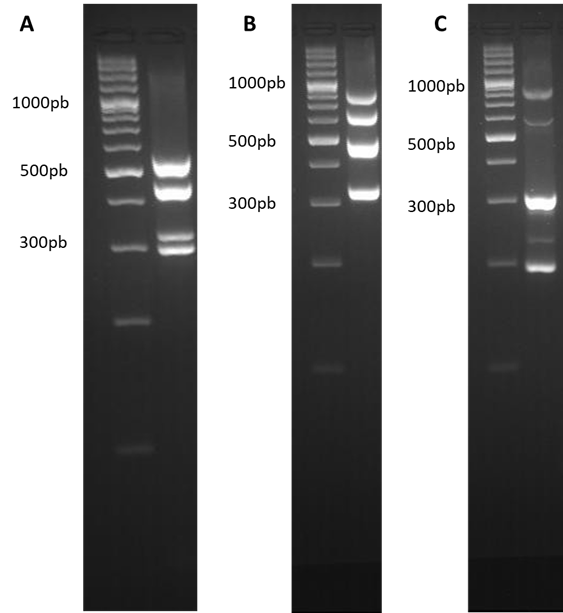
Figure 1 Multiplex PCR. Amplification bands for each of the multiplex PCR, (A) Multiplex 1, iroN 553 bp, hlyF 450 bp, Iss 332 bp and iutA 302 bp. (B) Multiplex 2, frz 843 bp, sitA 661 bp, vat 498 bp and KpsM 335 bp. (C) Multiplex 3, sopB 797 bp, sitD 571 bp, fimH 331 bp, uvrY 286 bp and pstB 198 bp. The gels were prepared at 4% agarose and a 100 bp weight marker was used.
The selected genes were amplified from the pool of control strains, and from the different isolates. For the multiplex PCR 1, a total of 37 isolates (78.7%) were positive for iroN, and 10 (21.3%) were negative, 38 isolates (80.9%) were positive for hlyF, and 9 isolates (19.1%) were negative, 3 isolates (6.4%) were positive for iss, and 44 (83.6%) were negative, and 36 isolates (76.6%) were positive for iutA, and 11 (23.4%) were negative. The profile of the bands generated can be seen in Fig. 2.
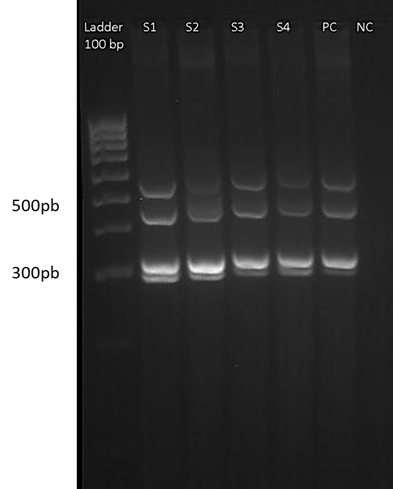
Figure 2 Multiplex PCR 1. The gel image shows one of the runs of multiplex 1, for 4 samples of isolates. From left to right: 100 bp weight marker, samples coded from S1 to S4, positive control (PC) and negative control (NC). The ladder serves as a reference to locate the position corresponding to the band of the amplicons of each of the genes (iroN, hlyF, Iss and iutA). The gel was prepared at 4% agarose.
In the multiplex PCR 2, a total of 23 isolates (48.9%) were positive for frz, and 24 (51.1%) were negative, 30 isolates (63.8%) were positive for vat, and 17 (36.2%) were negative, 35 isolates (74.5%) were positive for sitA, and 12 (25.5%) were negative, and 19 isolates (40.4%) were positive for KpsM, and 28 (59.6%) were negative. The profile of the bands generated can be seen in Fig. 3. In multiplex PCR 3, a total of 32 isolates (68.1%) were positive for sitD, and 15 (31.9%) were negative, all 47 isolates (100%) were positive for fimH, as well as for pstB, 32 isolates (68.1%) were positive for sopB, and 15 (31.9%) were negative, and finally 11 isolates (23.4%) were positive for uvrY, and 36 (76.5%) were negative.
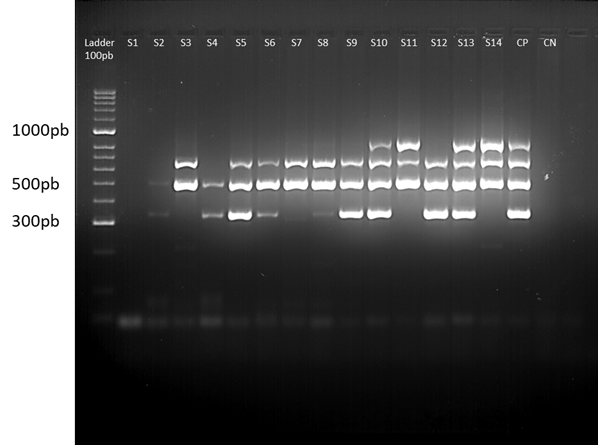
Figure 3 Multiplex PCR 2. The gel image shows one of the runs of multiplex 2, for 14 samples of isolates. From left to right: 100 bp weight marker, samples coded from S1 to S14, positive control (CP), negative control (CN) and 100 bp weight marker. The ladder serves as a reference to locate the position corresponding to the band of the amplicons of each of the genes (frz, sitA, vat and KpsM). The gel was prepared with 4% agarose.
The profile of the bands generated can be seen in Fig. 4. The isolates were classified into three profiles according to the virulence factors detected. In profile A, the isolates that had between 10 and 13 virulence factors. In profile B, those that presented between 5 and 9 virulence factors, and finally in profile C, those who had 4 or less virulence factors, which based on the genetic criteria for the pathogenicity were considered as the avian nonpathogenic Escherichia coli (non-APEC) strains (De Carli et al., 2015; Subedi et al., 2018). Out of 47 E. coli isolates, 39 (82.9%) isolates were found to be likely APEC strains due to the number of virulence factors, and 8 (17.02%) isolates were found to be non-APEC strains. For profile A, 20 isolates were found, and the maximum number of virulence factors detected was 12. For profile B, 19 isolates, and for profile C, 8 isolates, and the lowest number of virulence factors detected was 2. Among 47 E. coli strains, 1 strain contained twelve of the thirteen virulence genes evaluated, 13 strains contained eleven virulence genes, 6 strains contained ten virulence genes, 9 strains contained nine virulence genes, 5 strains contained eight virulence genes, 4 strains contained seven virulence genes, 1 strain contained six virulence genes, 1 strain contained 4 virulence genes, 2 strains contained 3 virulence genes, and 5 strains contained 2 virulence genes.
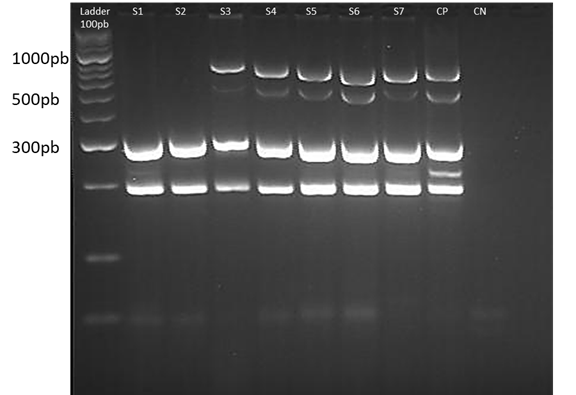
Figure 4 Multiplex PCR 3. The gel image shows one of the runs of multiplex 3, for 7 samples of isolates. From left to right: 100 bp weight marker, negative control (CN), positive control (CP) and samples coded from S1 to S7. The ladder serves as a reference to locate the position corresponding to the band of the amplicons of each of the genes (sopB, sitD, fimH, uvrY and pstB). The gel was prepared with 4% agarose.
The profile of the virulence factors of the vaccine strain was classified within profile C, with only 4 virulence factors (Table 2).
Table 2 List of analyzed samples, PCR result for each virulence factor [negative (-) and positive (+)] and virulence profile.
5. DISCUSSION
It has been calculated that there are over 26 billion chickens worldwide, with poultry constituting around 70% of all bird biomass on earth (Bar-On et al., 2018; Mageiros et al., 2021). According to the National Federation of Poultry Farmers (FENAVI) in Colombia, the poultry industry has grown considerably in the past decade, providing an anual output of more than one million tons of chicken meat during 2018, due to the implementation of hazard analysis and critical control point (HACCP) programs, in poultry slaughter establishments, as a voluntary measure to improve food safety. And even though Colombian farms meet international standards of environmental and animal welfare, which allow them to serve markets with special requirements (Ramírez-Hernández et al., 2017; FENAVI, 2021), the accurate data of prevalence of multi-antibiotic resistant strains or even pathogenic strains in Colombia is hardly documented, or at least reported, with exception of some initiatives.
The Colombian Integrated Surveillance Program for Antimicrobial Resistance (COIPARS) was established as a pilot project to monitor antimicrobial resistance among bacteria on poultry farms, slaughterhouses, and retail markets. This project helped to validate the methodology in the poultry chain in Colombia (Donado-Godoy et al., 2015).
In 2015 as a continuance of COIPARS, Donado-Godoy and collaborators established the baseline antimicrobial resistance patterns of Salmonella serovars, Escherichia coli, and Enterococcus spp., noticing that almost 98% of isolates tested were multidrug-resistant (Donado-Godoy et al., 2015). As a complement, Ramírez-Hernández and collaborators provided reference data for E. coli levels at various chicken processing steps in slaughterhouses in three representative regions of Colombia (Ramírez-Hernández et al., 2017). Finally, during 2017 Castellanos et al. reported that the resistance to extendedspectrum cephalosporins in E. coli from Colombian poultry samples, was mainly caused by Extended Spectrum Beta-Lactamases (ESBLs), and AmpC beta-lactamases, ESBL/AmpC genes including blaCMY-2 and blaSHV-12 (Castellanos et al., 2017).
Because of the lack of data and information gap regarding APEC in our country, and considering that some evidence suggests that most natural microbiota related to poultry production is not pathogenic to humans (del Río et al., 2007; J.-S. Kim & Kim, 2007), it is too early to know if APEC and ExPEC strains are phylogenetically related sharing some of the same virulence genes, and if there is a connection to antibiotics resistance, especially in APEC phylotypes which has been explored in other countries (Dziva & Stevens, 2008; Lutful Kabir, 2010; Borzi et al., 2018). All of this represents not only an important human, and animal health threat that requires persistent public health vigilance, but also a threat to the food chain production, as previously reported in some studies (Mitchell et al., 2015; Marazzato et al., 2020; Mageiros et al., 2021; Meena et al., 2021).
In this sense, the results of the three different multiplex PCRs have allowed us to detect many virulence factors in our E. coli isolates, which have shown a high prevalence in previous studies. The E. coli isolates in this study had at least two, and a máximum of 12 virulence genes, although the pathogenicity of APEC strains can be determined by the presence of at least five virulence genes (Wang et al., 2002; Rodriguez-Siek et al., 2005; Kemmett et al., 2013; De Carli et al., 2015; Subedi et al., 2018). And the frequency of each virulence factor when compared to previous reports worldwide, contributes to making a differentiation concerning the frequency of each factor in our isolates.
In the case of iroN, frequencies have been reported between 56 and 100% in APEC isolations (Rodriguez-Siek et al., 2005; van der Westhuizen & Bragg, 2012; Dissanayake et al., 2014; Varga et al., 2018). In our isolates, it was arround 80%, and as part of ExPEC strains, some studies had provided evidence that this iroN uptake system may also contribute to adherence and invasion during infection (Feldmann et al., 2007). Our study showed that the frequency of the iroN gene was closer to that found in the United States, Canada, and Nepal, than to those conducted in South Africa and Sir Lanka.
The hlyF gene is epidemiologically associated with virulent strains in APEC and human neonatal meningitis-associated E. coli. Recent studies have reported frequencies of 93.7% in Korea and 100% in Nepal. From the study in Nepal, they also managed to demonstrate within E. coli strains, a correlation between the presence of virulence factors and antibiotic resistance (Herren et al., 2006, p. 200; Y. B. Kim et al., 2020). In Brazil, the authors were able to detect the presence of this gene in 71.4% of the isolates (Silveira et al., 2016).
We obtaind a frequency of 80.9%, which showed to be higher than those reported in other SouthAmerican countries, but lower than those reported in Asia. That could be related to the absence is in those isolates with seven or fewer virulence factors, which might be related to less virulent strains. This virulence factor is directly involved in the production of outer membranevesicles, and their increased production was associated with the release of toxins during extraintestinal infection (Murase et al., 2016).
Regarding the iutA gene, the reported frequencies were very variable among the isolates, between 36%, and 100% (Gao et al., 2018; Subedi et al., 2018; Varga et al., 2018; Rahayuningtyas et al., 2020). In our study, the frequency was 76.6%, similar to iroN gene frequency (78.7%), both of these virulence genes encode siderophores and are essential for self-maintenance in the host body in APEC, and human ExPEC strains, contributing to their virulence (Rahayuningtyas et al., 2020). On the other hand, the Iss gene constantly has been reported in several studies with higher frequencies between 80 and 100% in the United States, Spain, Denmark, Nepal, Qatar, Korea, Canada, and Jordan (Kwon et al., 2008; Y.-W. Jeong et al., 2012; Solà-Ginés et al., 2015; El-Shaer et al., 2018; Subedi et al., 2018; Varga et al., 2018; Ibrahim et al., 2019; J. Jeong et al., 2021; Johar et al., 2021). Another study from the University of Florida reports a frequency of 75%, in which they also established that the presence of this gene could be associated with a health risk for humans (Stromberg et al., 2017). And even lower frequencies have been reported in isolates from Brazil (38.5%, 39.2%, and 49.5%) (Delicato et al., 2003; Silveira et al., 2016; Barbieri et al., 2021). However, our frequency was 6.4%, showing a huge difference from what was already reported.
The frz gene frequency has been reported in a study conducted in France with 15.1% (Schouler et al., 2012), and 22% in South Africa (van der Westhuizen & Bragg, 2012). However, the results of this study agree with those from Schouler et al., in 2012 with samples from France, Spain, and Belgium from chickens, turkeys, and ducks exhibiting clinical symptoms of different forms of colibacillosis, in which they found a prevalence of 53.4% compared to ours with 48.9% (Schouler et al., 2012). In the case of the fimH gene, as reported in Brazil, the United States, South Africa, and our study in Colombia, the frequency can vary from more than 80% to 100%, in our case fimH wasdetected in all the samples (Silveira et al., 2016; Stromberg et al., 2017; van der Westhuizen & Bragg, 2012). Related to the expression of these genes, the gene frz is chromosomally located and belongs to the frz operon, which encodes a phosphoenolpyruvate carbohydrate phosphotransferase system transporter and some enzymes involved in sugar metabolism. This has been proven to be relevant not only promoting bacterial fitness under stressful conditions but also seems to be involved in the cell Surface expression of F1 fimbriae (Rouquet et al., 2009).
The vat and sitA genes have been identified in both APEC and uropathogenic E. coli (UPEC) strains, both present in more than 60% of our strains. It has been demonstrated with in vitro, and ex vivo models that vacuolating autotransporter toxin induced cellular damage, vacuole formation, and urothelial barrier dysregulation of bladder epithelial cells (FENAVI, 2021). The frequency of the vat gene in Brazil was 13.2% (Silveira et al., 2016), while in Nepal reach out 89% (Subedi et al., 2018), and in England around 11.25% (Kemmett et al., 2013). In our isolates, the frequency was 63.8%, much higher than the one found in Brazil and England, but lower than in Nepal. For the sitA gene was reported a 20% in England (Kemmett et al., 2013), and with very similar values in Canada with 20.57% (Sabri et al., 2008). However, sitA was also found in over 85% of APEC, and as well human UPEC isolates, suggesting according to the authors, that it might enable APEC strains to cause extraintestinal disease in human beings (Rodriguez-Siek et al., 2005; Timothy et al., 2008). In recent studies of APEC isolates the most common gene identified was sitA, one from broiler and broiler breeder chickens in Ontario, Canada detected in 93% of the isolates (Varga et al., 2018), and another in broiler chickens in Jordan, with 97.4% (Ibrahim et al., 2019), our study shows a frequency of 74.5%. Our results showed that the gene frequency of KpsM, sitD, and pstB are higher than reported. The KpsM, pstB genes had the lowest prevalence, with only 2.2% in Zimbabwe (Mbanga & Nyararai, 2015).
In the case of the KpsM gene, a frequency of 10% in the United States (Stromberg et al., 2017), in Brazil of 14.3% (Silveira et al., 2016), and our isolates in Colombia were around 40%. The frequency of the sitD gene was 11% in Brazil (Sabri et al., 2008), 50% in Canada (Sabri et al., 2008), and 42.8% in South Africa (van der Westhuizen & Bragg, 2012). We reported a frequency of 68.1% in our isolates. And for the pstB gene, almost 86% in South Africa (van der Westhuizen & Bragg, 2012), and in our isolates, the frequency was 100%. It has been reported that the sopB and uvrY genes have not been detected in different avian fecal E. coli (AFEC) isolates from a variety of species of birds, such as turkeys, geese, and ducks (Dube & Mbanga, 2018). However, the gene sopB is not only highly prevalent among the Salmonella pathogenicity island genes and has a relevant impact on the pathogenesis and epidemiology of Salmonella infections in poultry (Ren et al., 2016; Ben Salem et al., 2017; Karacan Sever & Akan, 2019), but also was detected among the most prevalent virulence associated genes in APEC isolates, from confirmed cases of colibacillosis in chickens in Bulawayo, Zimbabwe, with a 20%, contrasting a 4.4% for the uvrY gene (Mbanga & Nyararai, 2015). In addition, from 10 Zimbabwean APEC isolates, the gene uvrY was detected in all the samples and the sopB gene in the 30% (van der Westhuizen & Bragg, 2012). In our isolates, the sopB gene was detected in 68.1% and the gene uvrY in 23.4% of them.
We were able to identify the presence of a group of virulence factors in bird clinical isolates of APEC in the Caloto region in Colombia, that share similarities as well as differences in both the frequency and the profile of virulence factors of those reported by other authors in other countries. As a particular trait pstB and fimH genes were present in the total of our isolates, and Iss gene had the lowest frequency, and the most common profile, was profile A, strains with between 10 and 13 factors of virulence.
CONCLUSIONS
According to published reports, the ColV plasmid has been considered an epidemiological marker of APEC, our study reported the presence of four of the five ColV plasmidencoded genes (IroN, hlyF, iss, and iutA), more than 80% of our strains showed one or a combination of at least 3 of these genes. In the case of iss despite the fact is considered as a candidate target of colibacillosis control procedures, and is strongly associated with APEC strains, it has been also detected in AFEC isolates from healthy birds (Nolan et al., 2002; Lynne et al., 2006; Goudarztalejerdi et al., 2020; Barbieri et al., 2021). Our results showed a very low frequency, that could be related to varying copy number of iss gene and hence its detection in our strains, or maybe related to individual genetic characteristics of the animals (Xu et al., 2018). Likewise, it has been shown that some E. coli strains isolated from colibacillosis cases, lacks important virulence genes without losing their pathotype and that there is no knowledge of a specific gene to be essential for the development of the extraintestinal infection in birds contributing to APEC virulence, and hence their relationship with human infection. According to this, and following the concept that pathogenicity is multifactorial and can arise from enriched commensal populations (Mageiros et al., 2021). We shouldn’t only be aware of the relationship between the presence of genes and E. coli virulence but also the effect of the sequence and transcription level, as well as the role of the intensity of the disease, risk factors, even environmental conditions (Delicato et al., 2003; Sadeghi Bonjar et al., 2017), potential antimicrobial resistance, and the frequency of virulence genes from isolates of healthy chickens (Barbieri et al., 2021). Which in our country, through more defined and effective molecular monitoring could end up in improved animal welfare and human food chain safety.














