INTRODUCTION
Maize, corn (Zea mays L.) an annual monoecious plant, which belongs to the Poaceae family, has a cylindrical stem, long and thick leaves, its height ranges from one to three meters, being a species that reproduces by cross-pollination, with feminine and masculine flowers located at different places of the plant (Sánchez & Pérez, 2014).
This species has evolved in different microenvironments, which has generated a great genetic variability; maize is native and was domesticated by the indigenous peoples in the center of Mexico, since about 10.000 years ago, being one of the main food resources for human consumption, it also has a great socio-economic and cultural importance (González Castro et al. 2013).
For centuries the activities of farmers, from the first agricultural societies to the present ones have allowed the selection and development of new varieties of maize, which are commonly known as Creole maize, which have adapted to the different ecological, agronomic, climatic and soil conditions, covering the nutritional and cultural requirements. Being subjected to important processes of genetic erosion, related to the introduction of hybrid maize and market demands (Tadeo et al. 2015). It is important to mention the fact that, in February 2007, the ICA decreed commercial plantings of three varieties of transgenic maize, namely: Roundup Ready maize (from Monsanto), Herculex I Bt maize (from Dupont) and Bt YieldGard MON 810 maize (from Monsanto), in the departments of Córdoba, Sucre, Huila and Tolima (Grupo Semillas, 2007); this unthinking and unilateral decision was made without studies that demonstrate the safety and convenience of these technologies for Colombia and its farmers, seriously affecting the native corn genetic diversity.
Lost of genetic variability lead to the loss of genetic and biological diversity, therefore, without genetic variability, a population cannot evolve in response to changes in environmental variables facing an increased risk of extinction (Gálvez et al. 2010). For this reason, molecular techniques have been used as an efficient tool for investigations of variability and genetic diversity of populations, being used in the protection, characterization and conservation of plants. One of the most recognized techniques is the use of microsatellites, this type of molecular markers based on DNA have been used to identify races and determine the genetic structure of various endemic lines of interest, microsatellites or short repeated sequences (STRs) have been widely used for the characterization of genetic variability and the description of genetic structure of maize populations, for having high reliability and automation (Bedoya et al. 2010).
At the international level, molecular markers have become important genetic indicators to determine genetic diversity and within these microsatellites are determinant because of their codominant character and their wide use in a wide range of crop species, including Maize (Elçi & Hançer, 2015). Also Salami et al. (2016), characterized accessions of maize through microsatellites in Benin, reporting a high polymophism and genetic differentiation among the accessions, information necessary to improve the production and conservation of Maize.
The objective of this work was to determine the genetic diversity of a population of Creole maize (Zea mays L.) by using STR-type molecular markers in Puerto Libertador Córdoba-Colombia in order to broaden knowledge about the genetic diversity of Creole maize and contribute to the approach of conservation strategies.
MATERIALS Y METHODS
Study area. The origin of the studied plant material was collected in farms of Puerto Libertador (7° 53’17’’ N 75° 40’18” W), in the department of Córdoba, Colombia.
Collection of samples. Samples of 30 maize accessions were taken in Puerto Libertador, deposited in resalable bags, where silica gel was added for dehydration; they were labeled and transported to the Genetics Laboratory of the University of Córdoba, for the corresponding analysis.
Extraction of genomic DNA. Fragments of dry foliar tissue were placed in a porcelain capsule with liquid nitrogen, each of the samples was macerated until a very fine powder was obtained and they were stored at -20 ° C until their later use. The extraction of the genomic DNA was carried out from 75mg of the macerated material, using the extraction and purification kit of Wizard® Genomic DNA Purification (PROMEGA).
Quantification of DNA. In order to evaluate the quality and to estimate the amount of DNA obtained in the extraction, a 1% agarose gel was made in a horizontal electrophoresis chamber (Multi Sub Choice Trio) and run at 100w for 40min. 5μl of DNA from each sample and 5μl of molecular weight marker with range 100-10000bp, GeneRuler 100pb Plus DNA Ladder (Thermo Scientific) were seeded. The concentration of the extracted DNA was estimated by comparing the bands of the molecular weight marker.
Polymerase chain reaction (PCR) and fragment analysis. The microsatellite markers (Table 1) were amplified by the PCR technique. The reaction mixture had a final volume of 25μL, which included 0.2μM of dNTPs, 1.5X of Buffer, 1.5μM of MgCl2, 0.5μM of each primer (Forward and reverse), 0.05U / μl of enzyme Taq polymerase, 3.33ng / μL of genomic DNA and sterile water to reach the final volume.
Table 1 Characteristics of the microsatellite primers of Zea mays L. (12), sequence of primers, allele number and allelic range (bp).
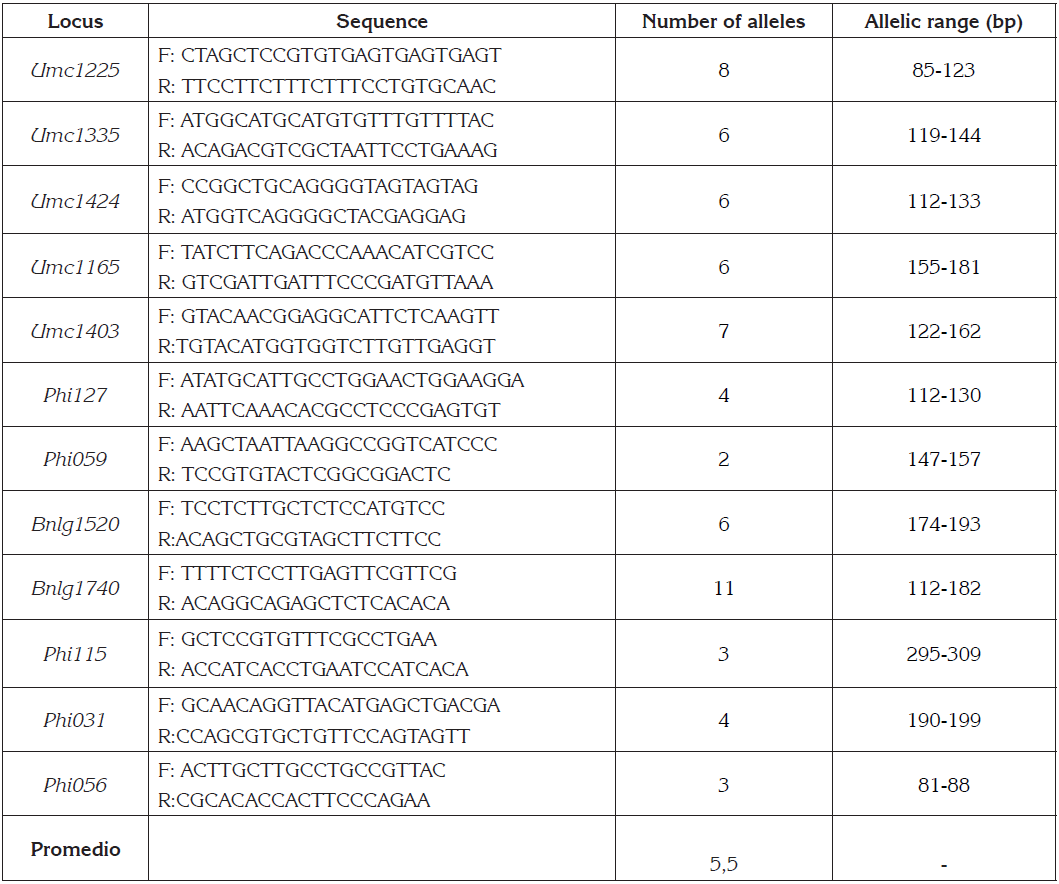
The PCR reaction was performed in a Bio-Rad T100 ™ thermocycler using the PCR Tochdown technique, which consisted of an initial denaturation phase of 95°C for 3 min, 12 cycles of: 30s denaturation at 95°C, 30s of annealing temperature from 62ºC to 57ºC reducing 1ºC every 2 cycles and an extension to 72ºC for 1min; 23 cycles with 30s of denaturation at 95°, 56ºC of annealing for 30s and extension at 72°C for 1min; and a final extension of 5min at 72°C.
The PCR products were separated by vertical electrophoresis in denatured (6 mol L-1 urea) 10% polyacrylamide gel (acrylamide: bisacrylamide, 29: 1) in a Mini-Protean II Biorad® camera (Applied Biosystems, Foster City, USA.) (Tsang et al. 1986). The bands were visualized by staining with silver nitrate (Qiu et al. 2012), using DNA ladder, where the range of the molecular weight marker used ranged between 50 and 500 bp. The gels were photographed with a CANON ELPH180 IS camera and the determination of the allelic size was carried out by the ImageJ program (Abràmoff et al. 2004) where, by pixelation of the amplified bands, the allelic sizes were determined using the software Past 3.14 (Hammer et al. 2001).
Statistical analysis. With the information obtained from the gels and using the software GENALEX 6 (Peakall and Smouse, 2006) it was calculated: number of alleles, effective number of alleles (Na), allelic frequencies, observed and expected heterozygosity values (Ho and He), fixation index (F) and Hardy-Weinberg equilibrium (HW). The PIC (Content of Polymorphic Information) of each microsatellite was determined by the CERVUS v. 3.0 Program (Kalinowski et al. 2007).
RESULTS AND DISCUSSION
In this study we found that the 12 analyzed molecular markers were polymorphic, presenting a total of 66 alleles, with a variation in the number of alleles between 2 (Phi59) and 11 (Bnlg1740) and the average number of alleles was 5.5 (Table 1).
For the 30 accessions of studied Creole maize, all the used microsatellite markers exhibited a high degree of polymorphism. The average allele number was 5.5, these values differ from those obtained by Fernandes et al. (2015), who reported average values of 5.7 and 2.8 alleles per locus respectively and similar to the result obtained by González Castro et al. (2013), who report values of 5.5.
The average of the effective number of alleles was 3,356, the marker with the highest effective number of alleles was Bnlg1740 with 6,844 and Phi059 had the lowest effective number of alleles with 1,839 (Table 2).
Table 2 Genetic parameters calculated in Zea mays L.
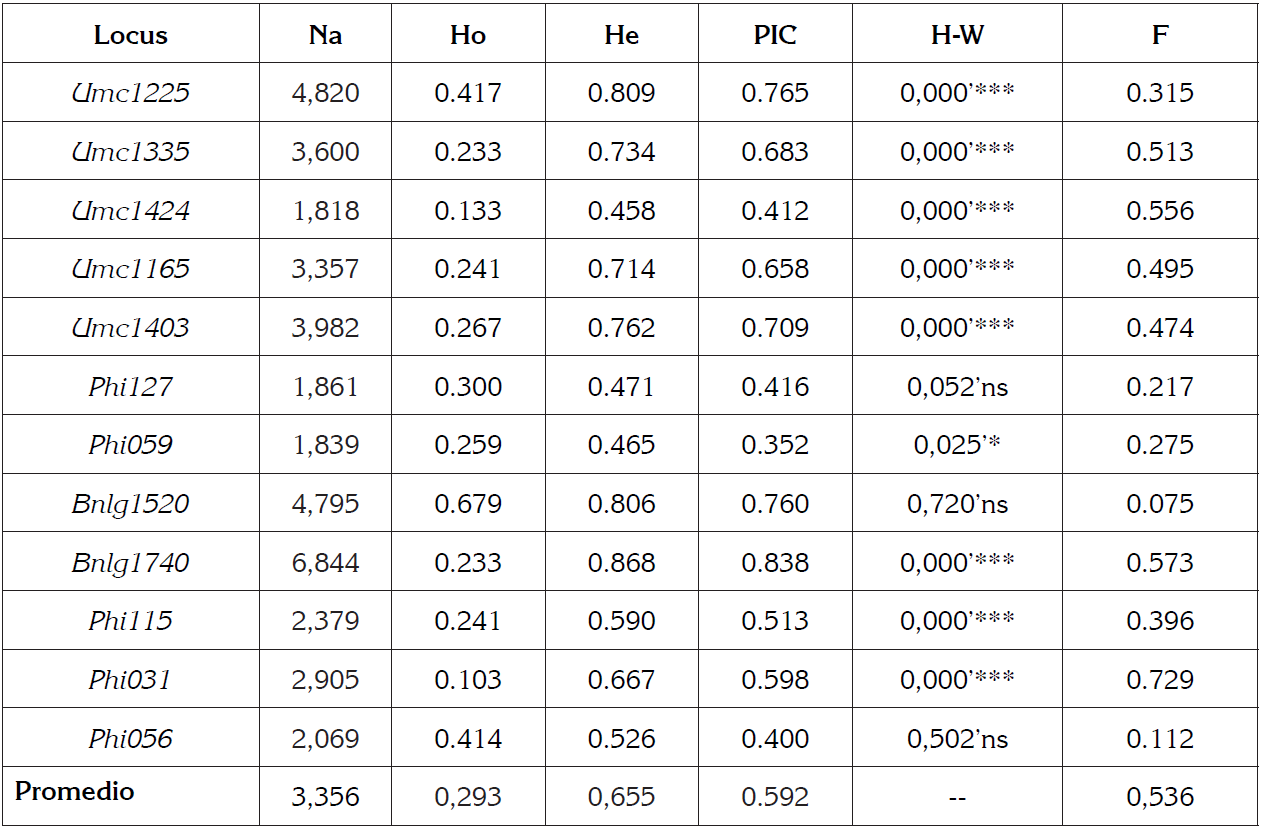
Na: Effective allele number; Ho: Heterozygosity observed, He: Heterozygosity expected; PIC: Polymorphic Information Content, HW: Hardy Weinberg equilibrium* P<0.05, ** P<0.01, *** P<0.001; F: fixation index.
The number of effective alleles (Na) for the population of Creole maize studied ranged between 1.81 and 6.84 with an average of 3.3, this result (Na = 3.3) was higher than that reported by Nyaligwa et al. (2015) (Na = 2.4) and lower than that obtained by Mora et al. (2013) (Na = 4.8) and Vivodík et al. (2017) (Na = 6.6).
The observed heterozygosity (Ho) average obtained for the 12 markers was 0.293, the marker Bnlg1520 obtained the highest value with 0.679 and the marker Phi31 obtained the lowest value with 0.103. The expected heterozygosity (He) average obtained was 0.655, the marker Bnlg1740 obtained the highest value with 0.868, and the marker Umc1424 obtained the lowest value with 0.458 (Table 2).
The expected heterozygosity (He) average of the 12 molecular markers evaluated was 0.65, revealing high genetic variability, as this is considered when values exceed 0.5 (Nei, 1978). This value is similar to that reported by Chen et al. (2016) (He = 0.690) and higher than other studies such as Salami et al. (2016) (He = 0.46).
The percentage of heterozygous individuals behaved below 50% for the heterozygosity observed average (Ho = 0.293), possibly by consanguineous mating or selection of seeds (Eguiarte et al. 2007). This value is lower than that reported by Bracco et al. (2013) (Ho = 0.341) and Chen et al. (2016) (Ho = 0.380).
The PIC obtained for the population varied between 0.352 (Phi059) and 0.838 (Bnlg1740), these values corresponding to the markers that presented the smallest and highest number of alleles. The average number of the PIC was 0.592 (Table 2). In this study, eight markers can be considered very informative (PIC> 0.5) and four moderately informative (PIC> 0.25).
Of the 12 markers used, 8 markers can be considered very informative (PIC> 0.5) to analyze the genetic variability of the Creole maize population and 4 moderately informative (PIC> 0.25), with the Bnlg1740 locus of greatest value (P = 0.838) and Phi059 (P = 0.352) the lowest value; these results demonstrate that microsatellite markers are appropriate for the evaluation of the genetic diversity of the species (Van et al. 2010). The average PIC (Polymorphic Information Content) obtained in the present study was 0.592, similar to that reported by Sharma et al. (2010), (PIC = 0.60) and Chen et al. (2016), (PIC = 0.52) and higher than that reported by Adeyemo et al. (2011) (PIC = 0.56), Fernandes et al. (2015) (PIC = 0.42), and lower than that reported by Ignjatovic-Micic et al. (2015) (PIC = 0.822) and Vivodík et al. (2017) (PIC= 0.814), this demonstrates the potential of microsatellites to detect differences between maize varieties (Al-Badeiry et al. 2014).
The population showed an absence of Hardy-Weinberg equilibrium (p <0.05) (Table 2). The markers Phi127 with 0.052, Bnlg1520 with 0.720, and Phi56 with 0.502 were found in Hardy-Weinberg equilibrium.
Of the 12 total markers, 9 showed imbalance, indicating that the population is not in Hardy-Weinberg equilibrium. This imbalance could be due to an excess of homozygotes, which can be attributed to inbreeding, bottlenecks, selection and domestication of plant material due to improper management of seeds by farmers (Bracco et al. 2013).
The average value of the fixation index (F) obtained for the 12 molecular markers evaluated was 0.536; the marker Bnlg1520 obtained the lowest value with 0.075 and the marker Phi31 obtained the highest value of F with 0.779, revealing an increase in the number of homozygotes with respect to the total population (Table 2).
The average fixation index obtained for the 12 molecular markers evaluated was 0.536, which may be due to external factors that intervene in the reproduction process of the plant, favoring cross-pollination or allogamy; this value is similar to that reported by Chen et al. (2016) (F = 0.45).
The results suggest that in order to maintain genetic diversity, it is necessary to design intervention mechanisms that not only stimulate the conservation of genetic material, but also contribute to improving the productive and economic results for farmers (Aguirre et al. 2010). allow access to the diversity of native varieties of the region, training in selection techniques and seed management to maintain valuable characteristics (Bellon et al. 2004).
Consideration should be given to prohibiting the planting of transgenic maize in the region and thus avoiding the loss of this important indigenous resource, which has been adapting to the environment for so many years, as well as the need for studies to determine the influence of the transgenic in the health of animals and humans
The microsatellites that provide the highest detection of polymorphism and therefore the highest informative quality are Umc1225, Umc1335, Umc 1165, Umc1403, Bnlg1520, Bnlg1740, Phi115 and Phi031 as they have the highest allelic richness and the highest polymorphic information content. Therefore, of the 12 markers evaluated, these six STR’s molecular markers are the indicators recommended to be included in later evaluations of genetic diversity for creole maize in Colombia.
The levels of expected heterozygosity in the present study indicate that the creole maize showed high genetic variability. The value of the fixation index (F) obtained revealed an increase in the number of homozygotes with respect to the total population.
It is necessary that genetic variability is further analyzed to increase knowledge about the genetic resources of the species and used for the design of strategies with characterization, production and conservation purposes.














