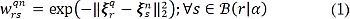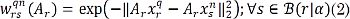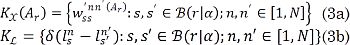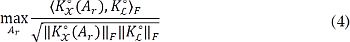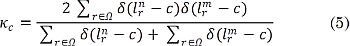Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
TecnoLógicas
versão impressa ISSN 0123-7799versão On-line ISSN 2256-5337
TecnoL. vol.20 no.39 Medellín maio/ago. 2017
Artículo de investigación/Research article
Multi-atlas label fusion by using supervised local weighting for brain image segmentation
Fusión de etiquetas basado en múltiples atlas usando ponderaciones locales supervisadas
D. Cárdenas-Peña1, E. Fernández2, José M. Ferrández-Vicente3 y G. Castellanos-Domínguez4
1 PhD in Engineering, Signal Processing and Recognition Group, Universidad Nacional de Colombia, Manizales-Colombia, dcardenasp@unal.edu.co
2 PhD in Bioengineering, CIBER BBN, Universidad Miguel Hernández, Elche-España, e.fernandez@umh.es
3 PhD in Informatics, DETCP, Universidad Politécnica de Cartagena, Murcia-España, jm.ferrandez@uptc.es
4 PhD in Engineering, Signal Processing and Recognition Group, Universidad Nacional de Colombia, Manizales-Colombia, cgcastellanosd@unal.edu.co
Fecha de recepción: 03 de abril de 2017/ Fecha de aceptación: 03 de mayo de 2017
Como citar / How to cite
D. Cárdenas-Peña, E. Fernández, J. M. Ferrández-Vicente y G. Castellanos-Domínguez, “Multi-atlas label fusion by using supervised local weighting for brain image segmentation”, TecnoLógicas, vol. 20, no. 39, 2017.
Abstract
The automatic segmentation of interest structures is devoted to the morphological analysis of brain magnetic resonance imaging volumes. It demands significant efforts due to its complicated shapes and since it lacks contrast between tissues and intersubject anatomical variability. One aspect that reduces the accuracy of the multi-atlas-based segmentation is the label fusion assumption of one-to-one correspondences between targets and atlas voxels. To improve the performance of brain image segmentation, label fusion approaches include spatial and intensity information by using voxel-wise weighted voting strategies. Although the weights are assessed for a predefined atlas set, they are not very efficient for labeling intricate structures since most tissue shapes are not uniformly distributed in the images. This paper proposes a methodology of voxel-wise feature extraction based on the linear combination of patch intensities. As far as we are concerned, this is the first attempt to locally learn the features by maximizing the centered kernel alignment function. Our methodology aims to build discriminative representations, deal with complex structures, and reduce the image artifacts. The result is an enhanced patch-based segmentation of brain images. For validation, the proposed brain image segmentation approach is compared against Bayesian-based and patch-wise label fusion on three different brain image datasets. In terms of the determined Dice similarity index, our proposal shows the highest segmentation accuracy (90.3% on average); it presents sufficient artifact robustness, and provides suitable repeatability of the segmentation results.
Keywords: Brain image segmentation, label fusion, multi-atlas segmentation.
Resumen
La segmentación automática de estructuras de interés en imágenes de resonancia magnética cerebral requiere esfuerzos significantes, debido a las formas complicadas, el bajo contraste y la variabilidad anatómica. Un aspecto que reduce el desempeño de la segmentación basada en múltiples atlas es la suposición de correspondencias uno-a-uno entre los voxeles objetivo y los del atlas. Para mejorar el desempeño de la segmentación, las metodologías de fusión de etiquetas incluyen información espacial y de intensidad a través de estrategias de votación ponderada a nivel de voxel. Aunque los pesos se calculan para un conjunto de atlas predefinido, estos no son muy eficientes en etiquetar estructuras intrincadas, ya que la mayoría de las formas de los tejidos no se distribuyen uniformemente en las imágenes. Este artículo propone una metodología de extracción de características a nivel de voxel basado en la combinación lineal de las intensidades de un parche. Hasta el momento, este es el primer intento de extraer características locales maximizando la función de alineamiento de kernel centralizado, buscando construir representaciones discriminativas, superar la complejidad de las estructuras, y reducir la influencia de los artefactos. Para validar los resultados, la estrategia de segmentación propuesta se compara contra la segmentación Bayesiana y la fusión de etiquetas basada en parches en tres bases de datos diferentes. Respecto del índice de similitud Dice, nuestra propuesta alcanza el más alto acierto (90.3% en promedio) con suficiente robusticidad ante los artefactos y respetabilidad apropiada.
Palabras clave: Segmentación de imágenes cerebrales, fusión de etiquetas, segmentación con múltiples atlas.
1. Introduction
Magnetic Resonance Imaging (MRI) is widely used for the identification of pathological brain changes [1], for building realistic head models [2], [3], and for brain tumor diagnosis [4], among other applications. Since the applications above rely on the features of anatomical structures of interest, brain image segmentation becomes an essential task, as it influences the outcome of the entire analysis [5]. This task is the process of tagging the voxels in the image with biologically meaningful labels. Nevertheless, the conventional manual annotation is a tedious and very time-consuming task. It highly depends on the skills of the expert, making its use impractical for most clinical applications [6]. On the other hand, automatic methods attempt to provide reliable results when applied to images acquired under different condi-tions. However, conditions like structural complexity, lack of contrast, and high anatomical variability make it hard to perform the labeling [7]. To cope with these limitations, automated segmentation algorithms incorporate a priori spatial information about the brain structures as pre-labeled images (termed atlases or templates). Consequently, atlas-guided segmentation encodes the relationship between the segmentation labels and image intensities of the atlases to further label the voxels of every unlabeled image [8]. The popularity of atlases stems from their widespread utility in guiding the segmentation process in areas of poor contrast and in helping to distinguish between tissues of similar intensities [9]. Since a single atlas may be not sufficient to cover the whole spectrum of variability within populations, multi-atlas strategies propose to combine the template images [10].
Related work
Multi-atlas segmentation strategies overcome the shape and size variability in brain structures by considering subject specific templates. Each template must be registered to the input volume so that the label fusion stage is carried out within the input coordinates [11]. One of these approaches selects the atlases with the greatest similarity to the input image, aiming to reduce the population average bias [12]. Then, the selected atlases feed segmentation models such as active shape patterns [13], appearance models [14], and probabilistic atlases [15]. These models highly rely on the accuracy supplied by the pairwise image alignment. In practice, such alignment is deteriorated when structures largely vary [16]. Although non-rigid transformations have been proposed to cope with this issue, registration is still very hard to perform in the presence of large structure deformations, as in the case of brain lesions or neurodegenerative disease [17].
To deal with the misregistration, the non-local methods identify all atlas-to-target agreements by using a template search strategy. As a result, the segmentation depends on the intensity similarity in a predefined neighborhood [10]. Recent approaches to non-local methods represent a voxel with intensity patches, but they differ in the selected fusion strategy. Thus, the basic methods estimate the labels by a weighted voting based on the similarity against neighboring patches [18]. Some examples are the weight computation that depends on the local registration performance [19] and the patch-based augmentation of the Expectation-Maximization with constrained search [20]. In more elaborate approaches, the input patch is reconstructed from the linear combination of patches from an a priori dictionary so that the mixing factors serve as the voting weights [21]. For instance, discriminative dictionary learning [22], local atlas selection [23], and contextual features [24] have been proposed to enhance the voting. Another approach suggests stratifying votes, depending on label purity and robust intensity statistics [25]. Recently, the sparse patch-based representation has been associated with tissue probability maps extracted from the registration stage [26]. Nonetheless, these techniques are limited by the patch-wise similarity, which is often globally handcrafted by the predefined features. This similarity reduces the segmentation performance for complexity structures [27].
In other approaches, the patch dictionary and its labels are used to learn an introduced classification function [28]. Also, the strengths of learning and weighted voting strategies join at the label fusion stage by the matrix completion method. Therefore, the partial label information obtained from the regions with least uncertainty allows to deal with the difficulty of space-varying labeling [29]. The probabilistic and patch-based approaches are combined as in [30]. In that case, they compute the target specific priors by a hierarchical scheme (accounting for global information) and a local patch search. In [31], a set of adaptive local priors is extracted from a set of training patches by using the local Markov random fields to model the local variations of appearance and shape. Also, a fuzzy c-means segmentation algorithm (in a CUDA accelerated version) enables to deal with local image artifacts [32]. Finally, [20] enhances the spatial priors required for the EM-based segmentation approach by applying a patch search algorithm. Nevertheless, the above methods demand a significant number of atlas voxels, reducing the benefit of the voting strategies [33]. Besides, there are regions with such uncertainty that very similar atlas patches may bear different labels (like the interfaces between two tissue structures), making it difficult to perform an accurate discrimination by using image similarity measures [34].
Our contribution
This paper introduces a multi-atlas weighted label fusion approach that fuses labels in a supervised learning scheme to improve the segmentation accuracy of brain MR images. In or-der to achieve this, we apply the knowledge about the neighborhood as well as the patch structure to be segmented. In accordance with the aforementioned, we have divided our contributions into several steps: Firstly, we adopt a novel methodology for feature extraction from the MRI voxels that is based on the linear combination of patch intensities, generalizing the convolution-like representations (e.g., gradients, Laplacians, and non-local means). Aiming to improve the accuracy of regions with intricate shapes, we adapt the features to the image location, assuming that structure complexity changes across the image. Secondly, we locally compute the linear projection by enhancing the similarity between the label and extracted feature distributions, building more discriminative representations and improving the interaction with image devices. To this end, we maximize the centered kernel alignment criterion. The latter assesses the correlation between a couple of kernel matrices [35]. Besides, we develop a neighborhood-wise procedure, providing more information about local properties of structures and avoiding the influence of small registration issues. Moreover, we fine-tuned the parameter by using two supervised criteria, namely: The radial relevance for patch radius selection, accounting for the impact of the voxels within patches on the attained projection; and the centered kernel alignment score, assessing the achieved correlation as a function of the neighborhood size. Both introduced criteria allow us to set up the algorithm off-line, instead of performing the segmentation for each parameter set. As a result, our proposal improves the segmentation accuracy and the performance under high artifact levels in comparison with baseline methods.
This paper is organized as follows: Section 2 describes multi-atlas segmentation as a standard non-local weighted voting label fusion. Section 2.2 introduces the mathematical description of the proposed supervised local feature learning. Section 3 describes the experiments that were carried out for tuning the algorithm parameters and the methodology for assessing the performance. In section 4 the achieved results for segmentation accuracy and repeatability are discussed. Finally, in Section 5, the concluding remarks and future research directions are presented.
2. Materials and methods
Let an input atlas dataset, , described by N triplets of pre-labeled images, with
, described by N triplets of pre-labeled images, with  as the intensity image, label image, and three-dimensional coordinates of the n-th atlas, respectively. Mn ∈ ℝ+ denotes the number of voxels in the n-th image. The intensity image, Xn={xnr ∈ ℝ}, collects the magnitude of the radio frequency signal generated at location r ∈ Ωn. The segmentation image, Ln={l nr ∈ 𝒞 }, stores the label assigned to each voxel. The label set, 𝒞 ={1,
,C}, designates the C considered anatomical partitions (tissues or structures of the brain). The set Ωn holds the coordinates, usually in millimeters, of each voxel within the volume in a three-dimensional space. The segmentation of an input image, Xq={xrq∈ ℝ :r ∈ Ωq}, designates a single label to each voxel coordinate. The multi-atlas-based segmentation registers the set A to the input volume, thus allowing to propagate the template labels to Ωq. Afterwards, all the labels are fused into a single class at coordinate r.
as the intensity image, label image, and three-dimensional coordinates of the n-th atlas, respectively. Mn ∈ ℝ+ denotes the number of voxels in the n-th image. The intensity image, Xn={xnr ∈ ℝ}, collects the magnitude of the radio frequency signal generated at location r ∈ Ωn. The segmentation image, Ln={l nr ∈ 𝒞 }, stores the label assigned to each voxel. The label set, 𝒞 ={1,
,C}, designates the C considered anatomical partitions (tissues or structures of the brain). The set Ωn holds the coordinates, usually in millimeters, of each voxel within the volume in a three-dimensional space. The segmentation of an input image, Xq={xrq∈ ℝ :r ∈ Ωq}, designates a single label to each voxel coordinate. The multi-atlas-based segmentation registers the set A to the input volume, thus allowing to propagate the template labels to Ωq. Afterwards, all the labels are fused into a single class at coordinate r.
The main image segmentation procedures are outlined as follows: i) Image registration computes a spatial transformation τn:Ωn → Ωq;r ↦ tn(r) maximizing the alignment between Xn and Xq, so that the atlases share the coordinates of the input volume. ii) Label propagation maps each n-th label image to the input coordinates through the transformation tn, yielding the label set 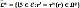 . iii) Label fusion combines all labels assigned to each voxel
. iii) Label fusion combines all labels assigned to each voxel 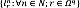 into a single label lrq ∈ 𝒞 , resulting in the segmented image
into a single label lrq ∈ 𝒞 , resulting in the segmented image 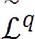 .
.
2.1 Feature-based label fusion within a-neighborhoods
The widely used multi-atlas label fusion builds a set of discriminative functions, noted as  , computing the level of membership to class c. Therefore, the labeling criterion for each voxel r ∈ Ωq is given by
, computing the level of membership to class c. Therefore, the labeling criterion for each voxel r ∈ Ωq is given by 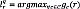 . In practice, label estimation degrades due to misregistration issues, low-frequency artifacts, and complicated shape structures. Within the locally weighted segmentation scheme, we introduce the following weighting factor, that varies along the space domain s, into 𝒢
. In practice, label estimation degrades due to misregistration issues, low-frequency artifacts, and complicated shape structures. Within the locally weighted segmentation scheme, we introduce the following weighting factor, that varies along the space domain s, into 𝒢
where ξ sn ∈ ℝ H is the H-dimensional feature representation of the s-th voxel in the n-th atlas, and  is the neighborhood centered at r, with a radius a ∈ ℝ +. Notations ∥·∥Ω and ∥·∥2 denote the norm along the three-dimensional coordinates Ω and the L2-norm, respectively. Then, we rewrite each c-th discriminating function in terms of the weighting factors as gc(r)=wrTδrc, where the vector wr={wrsnq ∈ ℝ +:n ∈ [1,N]} ∈ ℝ Ns holds Ns=NS weights computed for the S voxels in the a-neighborhood B (r|a), contributing to the labeling at location r.
is the neighborhood centered at r, with a radius a ∈ ℝ +. Notations ∥·∥Ω and ∥·∥2 denote the norm along the three-dimensional coordinates Ω and the L2-norm, respectively. Then, we rewrite each c-th discriminating function in terms of the weighting factors as gc(r)=wrTδrc, where the vector wr={wrsnq ∈ ℝ +:n ∈ [1,N]} ∈ ℝ Ns holds Ns=NS weights computed for the S voxels in the a-neighborhood B (r|a), contributing to the labeling at location r. 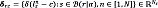 defines the vector of votes for the c-th label. Hence, the segmentation criterion now becomes lrq=argmax∀c∈CwrTSrc. As a result, the contribution of the voting voxels depends on their similarity to the features of the input voxel ξrq, contained in the corresponding voting weight wr.
defines the vector of votes for the c-th label. Hence, the segmentation criterion now becomes lrq=argmax∀c∈CwrTSrc. As a result, the contribution of the voting voxels depends on their similarity to the features of the input voxel ξrq, contained in the corresponding voting weight wr.
2.2 Supervised fusion weights based on centered kernel alignment
Aiming to include the local intensity information in the estimation of the voting weights wr, we propose to calculate the feature vector in (1) by using the linear projection ξsn= Ar xsn, where xsn={xtn: t∈ B(s|ß)} ∈ ℝ P denotes the vector containing all P voxel intensities within the patch centered at s with radius ß ∈ ℝ + termed the beta-patch, and the H×P-sized matrix Ar holds scalars Ar={ahpr∈ ℝ :h ∈ [1,H],p ∈ [1,P]}. Each of the scalars represents the contribution factor of the p-th voxel to build the h-th feature. Then, we compute the weights in (1) with the following kernel function:
Particularly, we looked for a projection matrix Ar allowing the weights in (2) to represent the similarity in both the feature and label spaces. To make the most of the available atlas information, the introduced supervised learning scheme arranges the subset of labeled voxels into two matrices: one holding the pairwise weighting factors and another one that accounts for label similarities. Both matrices are defined for the patch vectors within the a-neighborhood as follows:
Consequently, the more similar the matrices KX ∈ ℝ Ns×Ns and KL ∈ ℝ Ns×Ns, the more related they are to the extracted features and the labels. Moreover, the symmetry and positive definiteness properties of both, KX and KL, allow to find Ar by maximizing such similarity. The latter is measured in terms of the centered kernel alignment (CKA) criterion, as in [35]:
where K°=HKH, is the centered version of K, H=I-11T/Ns, and 1 is the all-ones vector with size Ns. Notations <·,·>F and ∥·∥F stand for the matrix Frobenius product and Frobenius norm, respectively. Therefore, the optimal matrix generates the most discriminative weighting factors regarding the input set of labeled voxels. The introduction of this supervised weight computation into the majority voting scheme is named Centered Kernel Alignment-based Label Fusion (CKA-LF).
3. Experimental setup
3.1 Database description
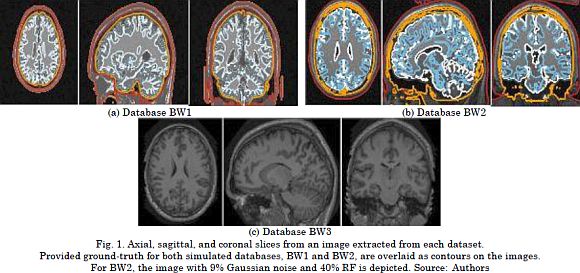
We validated the proposed CKA-LF by segmenting MRI volumes into five brain components, namely: Scalp (SC), skull (SK), cerebrospinal fluid (CSF), gray matter (GM), and white mat-ter (WM). The following three datasets are considered for assessing the segmentation per-formance (see Fig. 1 for sample images):
BrainWeb (BW1). This collection holds data generated by the BrainWeb MRI simulator. Phan-tom atlas subjects were 20 normal adults (ten females and ten males), ages 24 to 37 (Avg. 29.6 y/o). All MRI data were simulated by a 1.5 T Siemens Sonata Vision clinical scanner with a 30º flip angle, 22ms repeat time, 9.2ms echo time, and 1mm isotropic voxel size. This yields volumes of 256 x 256 x 181 voxels in size. Besides, a percentage noise level of 3% with no bias field was applied. The volumes in the BW1 collection are used to tune algorithm parameters, and their resulting segmentations are compared against the ground-truth labels.
BrainWeb (BW2). This collection contains 18 artificial images of the same subject. The images are generated by the BrainWeb MRI simulator for a T1-weighted contrast with a 308º flip angle, 18ms repeat time, and 10ms echo time to obtain 181 x 217 x 181-sized (1 x 1 x 1mm) volumes. The parameters for image artifacts are three different levels of the bias field (0, 20, 40% RF) and six levels of noise (0, 1, 3, 5, 7, 9%). The variety of noise parameters in this collection enables to evaluate the tuned algorithm for robustness by segmenting each BW2 image and using the BW1 images as the atlas dataset.
OASIS dataset. From the 416 subjects, we analyzed a collection of 20 normal anonymized real subjects. They were scanned twice in a 90-day period. In this subset, there are 12 fe-males and 8 males, ages 19 to 34. For each imaging session, a motion-corrected, co-registered average was extracted from three or four repetitions of the same T1-weighted structural protocol. The aim was to increase the signal-to-noise ratio. All the images were acquired using a 1.5-Tesla Vision scanner with 10º flip angle, 9.7ms repeat time, and 4ms echo time. As a result, we obtained 128 slices at a resolution of 1 x 1mm (256 x 256 pixels). In this paper, the OASIS dataset is considered for evaluating the repeatability of the results by comparing the resulting segmentations from the same subject.
3.2 Methodology description
In accordance with Section 2, brain tissue segmentation is divided into three main stages: Image registration, label propagation, and label fusion. For the first stage, the Advanced Normalization Tool (ANTs) spatially aligns the images and it is thoroughly evaluated for quan-titative morphometric analysis [36]. The rigid and deformable transformations were calculated by a quaternion-based mapping and the Elast elastic function. Thus we set the image metric to mutual information with 32-bins histograms. Deformable registration was performed at three sequential resolution levels: i) the coarsest alignment at 1/8 x Original space and 100 iterations, ii) the middle resolution at 1/4 x Original space and 50 iterations, and iii) the finest one at 1/2 x Original space and 25 iterations.
Lastly, the Gaussian regularization was carried out for the fixed scale σ=3 at each resolution level. In the second stage, the above-calculated deformations were applied to the label images by using a 0-th order interpolator. The goal was mapping the atlas labels to the input coordinates.
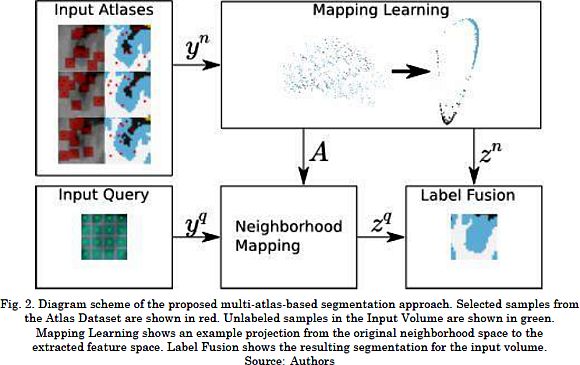
Since our contribution focuses on the third stage, we achieve target segmentation as follows. i) Patch representation describes atlas voxels, by using their labels and patches (red dots and patches in Atlas Dataset block of Fig. 2); and input voxels, by using their patches (green patches in Input Volume block of Fig. 2). ii) Feature learning estimates the matrix Ar from the neighbors of the input voxel in the atlas dataset by maximizing (4). iii) Feature extraction linearly projects patches onto the feature space by using the optimal matrix. iv) Label fusion computes voting weights by assessing the similarity of feature vectors following (1). The latter assigns the most voted structure as the label of output voxels.
3.3 Neighborhood and patch parameter tuning
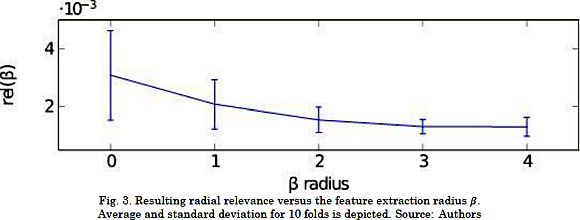
First, we adjusted the size of the neighborhoods and the patch as the most critical parameters of the proposed CKA-LF approach. Therefore, the assumed voxel representation by their appropriate ß-patches leads to the following issues: The larger the patch, the higher the computation cost; and the more remote the elements, the lesser their influence on the fusion label. According to this, we searched for the smallest ß with the largest alignment in the dependence of the centered alignment on the patch radius. To that end, we used the introduced radial relevance rel(ß)= 𝔼 {|ahp |:∥ρ ∥Ω=ß}, where notation 𝔼{·} is the averaging operator, ∥·∥Ω is the considered norm for the coordinates that we define as the L∞-norm, i.e. 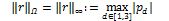 for evenly distributed voxels.
for evenly distributed voxels.
Since the proposed tuning of the patch radius requires the radial relevance, we computed the projection matrix Ar from a subset of Ns=104voxels that were randomly sampled from the BW1 collection. Fig. 3 displays the relevance values that were calculated by a 10-fold cross-validation strategy for values ß={0, ,4}. The results show that the closer the voxels are to the patch center, the larger their contribution is to the projection matrix. This behavior remains until the relevance reaches a steady value at ß=3. We consider the latter to be the optimal radius for the subsequent analysis.
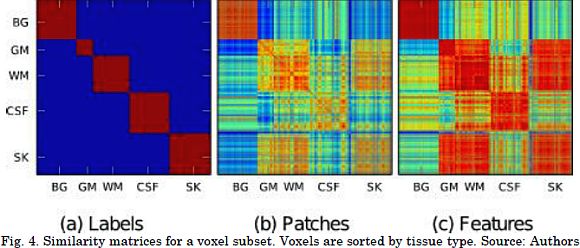
An example of the label similarity matrix, non-projected weights (i.e. Ar= I), and supervised weights (after CKA-based projection) is provided in Figs. 4a to 4c, respectively. All the values were computed for the optimal patch radius in the above sample subset. The voxels are displayed according to their tissue label. Fig. 4 shows that the supervised weights discriminate tissues better than the ones obtained from the patch vectors. This indicates a more accurate label fusion.
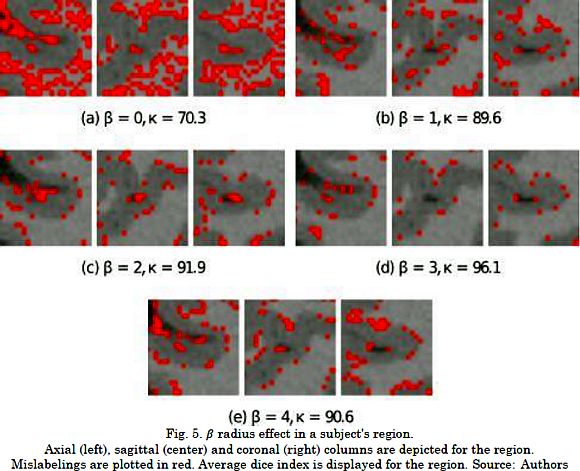
Fig. 5 presents the influence of the patch radius on the resulting labeling for a given region where the mislabeled pixels are marked in red. As it can be seen, ß=0 attains results in the lowest accuracy due to the lack of information to compute the required projection. On the other hand, the segmentation accuracy decreased for excessively large patches (see Fig. 5e). This is because CKA converged to an unsuitable maximum, the size of the projection matrix grew geometrically, and the patch distribution became more complex on large patches. Therefore, CKA benefits from incorporating spatial information into the voxel representation for particular patch radii.
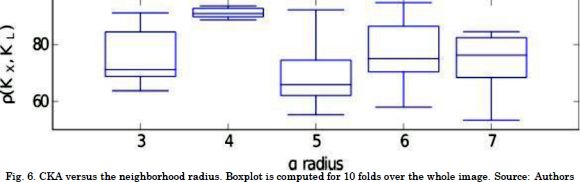
Then, we tuned the neighborhood radius by an exhaustive search for a and reached the best alignment between the reference and the intensity kernels. Fig. 6 presents the results of CKA scores for a≥ 3 in a 10-fold cross validation. The results prove that there is no statistical convergence for small alpha values due to the lack of neighbors. In contrast, for large neighborhoods, the linear projection cannot encode the label information correctly, since the samples are biased towards several structure shapes. This result derives from the fact that more complex than linear intensity relationships are induced. Particularly, a= 4 produces a robust alignment thanks to the trade-off between the number of neighbors and structure complexity. Consequently, a= 4 and ß=3 are the tuned algorithm parameters.
3.4 Performance segmentation measure
For evaluation purposes, CKA-LF is compared with two state-of-the-art methods: The Bayesian-based segmentation method (B-SEG) described in [8] and the patch-wise label fusion (Patch-LF) suggested by [37]. The former method does not require any parameter tuning. For the latter baseline algorithm, we set a=4 and ß=3 to compare both label fusion approaches in similar conditions. The performed segmentation is assessed in terms of the well-known Dice similarity index, Kc ∈ [0,1], defined by two label images, Ln and Lm, in the space domain Ω as in (5).
where Kc= 0 whenever the image regions of a given label do not overlap, and Kc= 1 if the tested regions become identical. Besides, we compute the global agreement between the performed segmentations by the average Dice index: 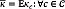
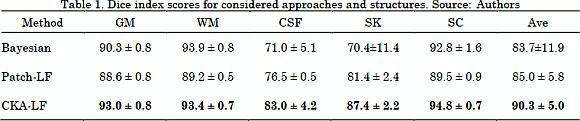
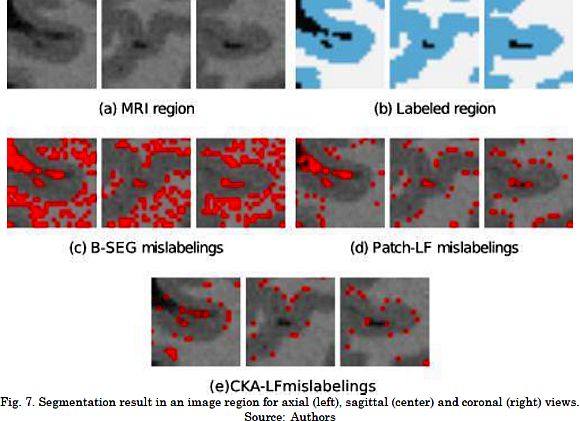
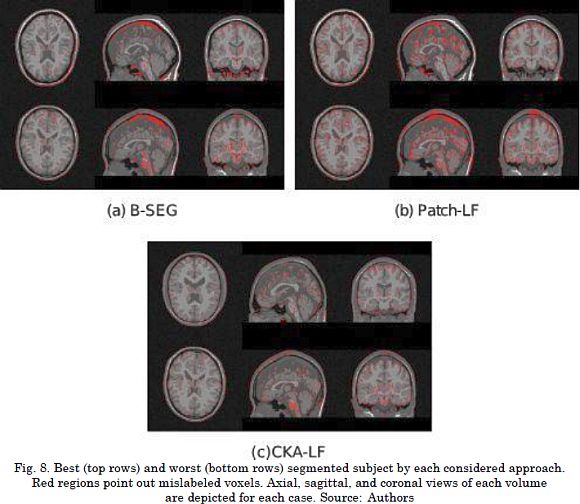
The results for the segmentation accuracy (obtained by the examined methods in BW1 dataset) are shown in Table 1 as the average and standard deviation values of kc. For the BW1 dataset, CKA-LF achieves the highest index (90.3 ± 5.0%), outperforming Patch-LF (85.0 ± 5.8%) and B-SEG (83.7 ± 11.9%). It should be noted that both testing methods of label fusion, Patch-LF and CKA-LF, provide more trustworthy results. This is because they achieve higher accuracies with lower deviations than the Bayesian-based approach. Besides, the use of voting schemes to segment the skull tissue improves the obtained accuracy in 20% while reducing the standard deviation. Fig. 7 and Fig. 8 are examples of the segmentation that was carried out on the BW1 dataset within an image region and for the whole image, respectively. Qualitative results prove that supervised label fusion yields the lowest number of mislabeling errors.
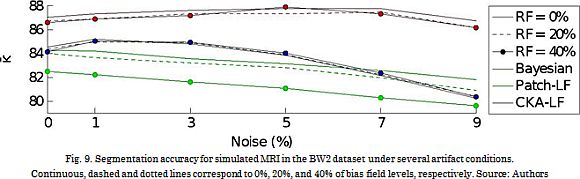
Further, the artifact robustness is assessed in the BW2 dataset for 6 noise levels and three levels of the bias field. The resulting performance curves (see Fig. 9) show that B-SEG suitably deals with low noise levels, while Patch-LF performs the worst. However, the proposed CKA-LF properly copes with both kinds of considered artifacts: noise and inhomogeneity
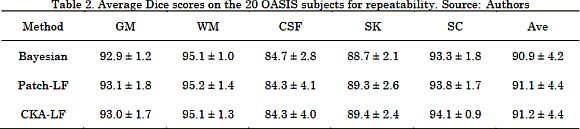
Finally, we validated the repeatability of the algorithms on the OASIS dataset. To this end, we measured the Dice index between the segmented images of the same subject. Therefore, a large k means that the algorithm better replicates the performance results. As shown in Table 2, each one of the tested approaches (B-SEG, Patch-LF, CKA-LF) achieved Dice index values over 90%. This implies that the algorithms can replicate the segmentation regardless of the input subject volume. Besides, the difference in the average Dice is lower than 1%, and the standard deviations are higher than 1%, yielding a statistically similar performance among approaches. As a result, the proposed CKA-LF method increases the segmentation quality with the additional benefit of reaching values of repeatability that are similar to the state-of-the-art.
4. Discussion
The new method of patch-based label fusion is proposed to improve automatic labeling of brain structures while dealing with low boundary contrast. CKA-LF benefits from the information provided by the label distribution of the atlases to locally learn the shapes. From the previously described validation, the following aspects are relevant in developing the CKA-LF method:
First, the voting function attempts to measure the pairwise similarity between the linearly extracted feature vectors. The feature extraction is carried out under a supervised learning scheme that places similarly labeled voxels closer. The centered kernel alignment criterion is introduced to quantify the similarity between features and labels. Thus, maximizing the CKA criterion and linearly projecting patches (see Fig. 4) result in a voting function that increases the class discrimination.
Second, patch and neighborhood radii strongly influence the estimation of the mapping function. Specifically, the former parameter determines the projection domain and the intensity variability inside each patch. The latter parameter establishes the number of available samples for estimating the projection matrix and the patch distribution within the neighborhood. This enables to cope with small registration issues. Therefore, the lack of patch information in the computed projection decreases the achievable accuracy of label fusion, when ß → 0, as seen in Fig. 5a . The performance also worsens for the very large patches (see Fig. 5e) due to the geometrically growing size of the resulting projection matrix and the complex distribution of the patch vectors. The radial relevance (that measures the influence of the radii on the projection) is introduced to tune ß. As seen in Fig. 3 , the relevance monotonically decreases as the distance grows. This means that the closer the voxels are to the patch center, the more they contribute to build the features. In the case of the a-neighborhoods, small radii provide more robustness to low-frequency artifacts due to the lack of patches; thus poorly estimating Ar (Fig. 6 ). By contrast, large a values produce more patches and increase the shape variability. Therefore, the projection matrix calculation is puzzled. Consequently, we balance the number of samples and the modeling complexity by maximizing the CKA regarding a.
We compare the proposed CKA-LF with the baseline Bayesian-based segmentation and with the Patch-wise label fusion methods in terms of their performed accuracy, robustness of artifacts, and algorithm repeatability. Regarding the estimated ¯? index, the proposed CKA-LF outperforms the other approaches in comparison (Table 1) and it has the additional benefit of lower accuracy deviations. Particularly, the cerebrospinal fluid and the skull tissues pose a major challenge for segmentation because of the few labeled samples. For these structures, CKA-LF highlights the contrast between both neighboring structures by incorporated projection. For the gray and white matter, CKA-LF learns their intricate shapes from the patch features and yields an accuracy boost of 5% compared to baselines. Regarding the qualitative results, Fig. 7 and 8, mostly locate mislabelings on the structure boundaries due to the lack of contrast between tissues. In terms of robustness, the validation in the BW2 dataset proves that enhancing the correlation between patches allows to better deal with noise and bias. It also increases the overall segmentation performance.
Lastly, all three compared approaches appropriately reproduce the outcome segmentation of the same subject in every dataset image (Table 2). Thus, the difference between the estimated K and K indexes is less than 1% with 1-4% of deviation. Although the reached k index is statistically equivalent for all the algorithms, the repeatability scores for cerebrospinal fluid and skull are lower than 90%. This result may be explained since both tissue boundaries change from scan to scan as a consequence of the limited image resolution and the reduced thickness of the examined tissue structures.
5. Conclusions
This research proposes a new multi-atlas weighted label fusion approach for brain image segmentation. This method makes the most of a more elaborated fusing procedure by incorporating the knowledge of the voxel neighborhood, as well as the patch structure of the considered tissues. For this purpose, all image patches are projected onto a discriminating space that maximizes the similarity between the labels and the feature vectors by using the introduced centered kernel alignment criterion. Besides, the adopted neighborhood-wise analysis enables to account more useful information on tissue structure localities, in order to avoid the influence of small registration issues on the input image. Therefore, the proposed CKA-LF effectively learns local structure shapes and reduces the artifact influence. Furthermore, we obtained a better accuracy under high artifact levels when compared to state-of-the-art methods for brain tissue segmentation (Bayesian-based segmentation and patch-wise label fusion). It is worth noting that CKA-LF performs all of the above with adequate repeatability scores. As future research, we propose the use of supervised local feature extraction for unified schemes involving registration, template selection, dictionary learning [38], and further studies on segmentation. Additionally, the extension of this paper could be focused on the segmentation of other structures, such as basal ganglia, and other imaging modalities, such as computed tomography.
6. Acknowledgments
This work was supported by the research Project 111974454838 funded by COLCIENCIAS.
References
[1] E. E. Bron, M. Smits, W. M. van der Flier, H. Vrenken, F. Barkhof, P. Scheltens, J. M. Papma, R. M. E. Steketee, C. Méndez Orellana, R. Meijboom, M. Pinto, J. R. Meireles, C. Garrett, A. J. Bastos-Leite, A. Abdulkadir, O. Ronneberger, N. Amoroso, R. Bellotti, D. Cárdenas-Peña, A. M. Álvarez-Meza, C. V. Dolph, K. M. Iftekharuddin, S. F. Eskildsen, P. Coupé, V. S. Fonov, K. Franke, C. Gaser, C. Ledig, R. Guerrero, T. Tong, K. R. Gray, E. Moradi, J. Tohka, A. Routier, S. Durrleman, A. Sarica, G. Di Fatta, F. Sensi, A. Chincarini, G. M. Smith, Z. V. Stoyanov, L. Sørensen, M. Nielsen, S. Tangaro, P. Inglese, C. Wachinger, M. Reuter, J. C. van Swieten, W. J. Niessen, and S. Klein, “Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: The CADDementia challenge,” Neuroimage, vol. 111, pp. 562-579, May 2015. [ Links ]
[2] J. D. Martínez-Vargas, G. Strobbe, K. Vonck, P. van Mierlo, and G. Castellanos-Dominguez, “Improved Localization of Seizure Onset Zones Using Spatiotemporal Constraints and Time-Varying Source Connectivity,” Front. Neurosci., vol. 11, p. 156, Apr. 2017. [ Links ]
[3] P. A. Valdés-Hernández, N. von Ellenrieder, A. Ojeda-González, S. Kochen, Y. Alemán-Gómez, C. Muravchik, and P. A. Valdés-Sosa, “Approximate average head models for EEG source imaging.,” J. Neurosci. Methods, vol. 185, no. 1, pp. 125-32, Dec. 2009. [ Links ]
[4] S. Pereira, A. Pinto, V. Alves, and C. A. Silva, “Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images,” IEEE Trans. Med. Imaging, vol. 35, no. 5, pp. 1240-1251, May 2016. [ Links ]
[5] R. Magalhães, P. Marques, J. Soares, V. Alves, and N. Sousa, “The Impact of Normalization and Segmentation on Resting-State Brain Networks,” Brain Connect., vol. 5, no. 3, pp. 166-176, Apr. 2015. [ Links ]
[6] J. Ahdidan, C. A. Raji, E. A. DeYoe, J. Mathis, K. Ø. Noe, J. Rimestad, T. K. Kjeldsen, J. Mosegaard, J. T. Becker, and O. Lopez, “Quantitative Neuroimaging Software for Clinical Assessment of Hippocampal Volumes on MR Imaging,” J. Alzheimer's Dis., vol. 49, no. 3, pp. 723-732, Oct. 2015. [ Links ]
[7] M. Ganzetti, N. Wenderoth, and D. Mantini, “Quantitative Evaluation of Intensity Inhomogeneity Correction Methods for Structural MR Brain Images,” Neuroinformatics, vol. 14, no. 1, pp. 5-21, Jan. 2016. [ Links ]
[8] J. Ashburner and K. J. Friston, “Unified segmentation,” Neuroimage, vol. 26, no. 3, pp. 839-851, Jul. 2005. [ Links ]
[9] P. F. Raudaschl, P. Zaffino, G. C. Sharp, M. F. Spadea, A. Chen, B. M. Dawant, T. Albrecht, T. Gass, C. Langguth, M. Lüthi, F. Jung, O. Knapp, S. Wesarg, R. Mannion-Haworth, M. Bowes, A. Ashman, G. Guillard, A. Brett, G. Vincent, M. Orbes-Arteaga, D. Cárdenas-Peña, G. Castellanos-Dominguez, N. Aghdasi, Y. Li, A. Berens, K. Moe, B. Hannaford, R. Schubert, and K. D. Fritscher, “Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015.,” Med. Phys., vol. 44, no. 5, pp. 2020-2036, May 2017. [ Links ]
[10] J. E. Iglesias and M. R. Sabuncu, “Multi-atlas segmentation of biomedical images: A survey,” Med. Image Anal., vol. 24, no. 1, pp. 205-219, Aug. 2015. [ Links ]
[11] J. V Manjón and P. Coupé, “volBrain: An Online MRI Brain Volumetry System.,” Front. Neuroinform., vol. 10, p. 30, Jul. 2016. [ Links ]
[12] P. Aljabar, R. A. Heckemann, A. Hammers, J. V Hajnal, and D. Rueckert, “Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy.,” Neuroimage, vol. 46, no. 3, pp. 726-738, Jul. 2009. [ Links ]
[13] F. M. Sukno, S. Ordas, C. Butakoff, S. Cruz, and A. F. Frangi, “Active Shape Models with Invariant Optimal Features: Application to Facial Analysis,” IEEE Trans. Pattern Anal. Mach. Intell., vol. 29, no. 7, pp. 1105-1117, Jul. 2007. [ Links ]
[14] B. Patenaude, S. M. Smith, D. N. Kennedy, and M. Jenkinson, “A Bayesian model of shape and appearance for subcortical brain segmentation,” Neuroimage, vol. 56, no. 3, pp. 907-922, Jun. 2011. [ Links ]
[15] C. Chu, M. Oda, T. Kitasaka, K. Misawa, M. Fujiwara, Y. Hayashi, Y. Nimura, D. Rueckert, and K. Mori, “Multi-organ Segmentation Based on Spatially-Divided Probabilistic Atlas from 3D Abdominal CT Images,” in MICCAI 2013: Medical Image Computing and Computer-Assisted Intervention - MICCAI 2013, Springer, Berlin, Heidelberg, 2013, pp. 165-172. [ Links ]
[16] F. E.-Z. A. El-Gamal, M. Elmogy, and A. Atwan, “Current trends in medical image registration and fusion,” Egypt. Informatics J., vol. 17, no. 1, pp. 99-124, Mar. 2016. [ Links ]
[17] I. Despotovic, B. Goossens, and W. Philips, “MRI Segmentation of the Human Brain: Challenges, Methods, and Applications,” Comput. Math. Methods Med., vol. 2015, pp. 1-23, 2015. [ Links ]
[18] F. Rousseau, P. A. Habas, and C. Studholme, “A Supervised Patch-Based Approach for Human Brain Labeling,” IEEE Trans. Med. Imaging, vol. 30, no. 10, pp. 1852-1862, Oct. 2011. [ Links ]
[19] I. Isgum, M. Staring, A. Rutten, M. Prokop, M. A. Viergever, and B. Van Ginneken, “Multi-atlas-based segmentation with local decision fusion-application to cardiac and aortic segmentation in CT scans,” IEEE Trans. Med. Imaging, vol. 28, no. 7, pp. 1000-1010, 2009. [ Links ]
[20] M. Liu, A. Kitsch, S. Miller, V. Chau, K. Poskitt, F. Rousseau, D. Shaw, and C. Studholme, “Patch-based augmentation of Expectation-Maximization for brain MRI tissue segmentation at arbitrary age after premature birth,” Neuroimage, vol. 127, pp. 387-408, Feb. 2016. [ Links ]
[21] D. Zhang, Q. Guo, G. Wu, and D. Shen, “Sparse Patch-Based Label Fusion for Multi-Atlas Segmentation,” in Multimodal Brain Image Analysis, vol. 7509, P.-T. Yap, T. Liu, D. Shen, C.-F. Westin, and L. Shen, Eds. Springer Berlin Heidelberg, 2012, pp. 94-102. [ Links ]
[22] T. Tong, R. Wolz, P. Coupé, J. V Hajnal, and D. Rueckert, “Segmentation of MR images via discriminative dictionary learning and sparse coding: Application to hippocampus labeling,” Neuroimage, vol. 76, pp. 11-23, [ Links ]
[23] T. Tong, R. Wolz, Z. Wang, Q. Gao, K. Misawa, M. Fujiwara, K. Mori, J. V Hajnal, and D. Rueckert, “Discriminative dictionary learning for abdominal multi-organ segmentation.,” Med. Image Anal., vol. 23, no. 1, pp. 92-104, Jul. 2015. [ Links ]
[24] W. Bai, W. Shi, C. Ledig, and D. Rueckert, “Multi-atlas segmentation with augmented features for cardiac MR images,” Med. Image Anal., vol. 19, no. 1, pp. 98-109, Jan. 2015. [ Links ]
[25] N. Cordier, H. Delingette, and N. Ayache, “A Patch-Based Approach for the Segmentation of Pathologies: Application to Glioma Labelling,” IEEE Trans. Med. Imaging, vol. 35, no. 4, pp. 1066-1076, Apr. 2016. [ Links ]
[26] M. Yan, H. Liu, X. Xu, E. Song, Y. Qian, N. Pan, R. Jin, L. Jin, S. Cheng, and C.-C. Hung, “An improved label fusion approach with sparse patch-based representation for MRI brain image segmentation,” Int. J. Imaging Syst. Technol., vol. 27, no. 1, pp. 23-32, Mar. 2017. [ Links ]
[27]G. Ma, Y. Gao, G. Wu, L. Wu, and D. Shen, “Atlas-Guided Multi-channel Forest Learning for Human Brain Labeling,” in Medical Computer Vision: Algorithms for Big Data, vol. 8848, B. Menze, G. Langs, A. Montillo, M. Kelm, H. Müller, S. Zhang, W. Cai, and D. Metaxas, Eds. Cham: Springer International Publishing, 2014, pp. 97-104. [ Links ]
[28] Y. Hao, T. Wang, X. Zhang, Y. Duan, C. Yu, T. Jiang, and Y. Fan, “Local label learning (LLL) for subcortical structure segmentation: Application to hippocampus segmentation,” Hum. Brain Mapp., vol. 35, no. 6, pp. 2674-2697, Jun. 2014. [ Links ]
[29] G. Sanroma, G. Wu, Y. Gao, K.-H. Thung, Y. Guo, and D. Shen, “A transversal approach for patch-based label fusion via matrix completion,” Med. Image Anal., vol. 24, no. 1, pp. 135-148, 2015. [ Links ].
[30] R. Wolz, C. Chu, K. Misawa, M. Fujiwara, K. Mori, and D. Rueckert, “Automated Abdominal Multi-Organ Segmentation With Subject-Specific Atlas Generation,” IEEE Trans. Med. Imaging, vol. 32, no. 9, pp. 1723-1730, Sep. 2013. [ Links ].
[31] S. Lee, S. H. Park, H. Shim, I. D. Yun, and S. U. Lee, “Optimization of local shape and appearance probabilities for segmentation of knee cartilage in 3-D MR images,” Comput. Vis. Image Underst., vol. 115, no. 12, pp. 1710-1720, Dec. 2011. [ Links ]
[32] C. Feng, D. Zhao, and M. Huang, “Image segmentation using CUDA accelerated non-local means denoising and bias correction embedded fuzzy c-means (BCEFCM),” Signal Processing, vol. 122, pp. 164-189, May 2016. [ Links ]
[33] O. V Senyukova and A. Y. Zubov, “Full anatomical labeling of magnetic resonance images of human brain by registration with multiple atlases,” Program. Comput. Softw., vol. 42, no. 6, pp. 356-360, Nov. 2016. [ Links ].
[34] G. Sanroma, O. M. Benkarim, G. Piella, G. Wu, X. Zhu, D. Shen, and M. Á. G. Ballester, “Discriminative Dimensionality Reduction for Patch-Based Label Fusion,” in Machine Learning Meets Medical Imaging, Springer, Cham, 2015, pp. 94-103. [ Links ].
[35] C. Cortes, M. Mohri, and A. Rostamizadeh, “Algorithms for Learning Kernels Based on Centered Alignment,” J. Mach. Learn., vol. 13, pp. 795-828, Mar. 2012. [ Links ]
[36] B. B. Avants, N. J. Tustison, G. Song, P. A. Cook, A. Klein, and J. C. Gee, “A reproducible evaluation of ANTs similarity metric performance in brain image registration.,” Neuroimage, vol. 54, no. 3, pp. 2033-44, Feb. 2011. [ Links ]
[37] P. Coupé, J. V Manjón, V. Fonov, J. Pruessner, M. Robles, and D. L. Collins, “Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation.,” Neuroimage, vol. 54, no. 2, pp. 940-954, Jan. 2011. [ Links ]
[38] S. Roy, A. Carass, J. L. Prince, and D. L. Pham, “Subject Specific Sparse Dictionary Learning for Atlas Based Brain MRI Segmentation,” in Mach Learn Med Imaging, vol. 8679, 2014, pp. 248-255. [ Links ]