1. INTRODUCTION
At the beginning of this century, fossil fuels accounted for approximately 80% of the global energy aimed at satisfying the world’s energy consumption. Subsequently, inappropriate handling of these materials and their waste has created a serious problem worldwide due to the pollution of the soil and water with crude oil and by-products [1].
A wide variety of environments could be contaminated with hydrocarbons. When the pollution occurs in soil, the hydrocarbons prevent gas exchange with the atmosphere, causing physicochemical changes like abnormal evaporation and penetration [2].
These compounds drastically change the conditions of salinity, causing the destruction of the tertiary structure of proteins, denaturation of enzymes, and dehydration of the cell, which is lethal to many organisms [2]. When the contamination occurs in water, a layer is generated by oil spills given the difference in density and thus preventing the entry of light and the gas exchange. Water-soluble compounds mix with water, which directly affects niches and ecosystems [2].
Due to the high toxicity of phenolic compounds, the Environmental Protection Agency of the United States and the European Union list them as dangerous pollutants [3]. The presence of phenols in the environment is an important risk for the aquatic biota, even at low concentrations [4].
Given the aforementioned problems, it is necessary to establish and develop technologies for environmental remediation. Most toxic compounds can be removed from environmental and industrial effluents by means of physicochemical methods; ozonation, activated carbon adsorption, chemical oxidation, Fenton’s reagent, UV, and hydrogen peroxide are some of them [2]. The disadvantages of these treatments include their complexity, costs, and the fact that they can produce equally dangerous byproducts [4]. For these reasons, exploring the biodiversity to find a solution became relevant, and bioremediation appears to be an interesting alternative. It involves the use of some species that, given their metabolic characteristics, can act in multiple environments neutralizing toxic substances or making them harmless for the environment or human health [2].
A wide variety of microorganisms have a huge biotechnological potential to treat polluted wastewater and soil [5]. Specifically, consortia of bacteria and microalgae have proven to be more efficient in the detoxification of organic and inorganic pollutants, as well as the removal of nutrients from wastewater, than the individual microorganisms [5]. Microalgal photosynthesis provides oxygen, a key electron acceptor to the pollutant-degrading heterotrophic bacteria. In turn, bacteria support the photoautotrophic growth of their partners by providing carbon dioxide and other stimulatory means for continuing the photosynthetic cycle [6].
Interactions between autotrophic algae and heterotrophic bacteria can be cooperative or competitive. Certain bacteria are often accompanied by microalgae, even under the laboratory conditions of unialgal cultures [6]. This close contact between aerobic bacteria and microalgae has been reported between Chlorella sp. and Pseudomonas sp. [6], and between Chlorella sp. and Stenotrophomonas maltophilia [7]. The stimulatory growth of both microorganisms has been observed in such consortia; furthermore, several compounds produced by the microalgae can support a faster better growth of the bacteria [6]. Microalgae produce sheaths (of trilaminar organization) which consist of carbohydrate, protein, and metal cations that are related to the formation of algal cell aggregation, in which bacteria are associated by indirect adhesion as bacterial symbionts on the sheath and by direct adhesion over the algal cell surface., As a result, the diffusion distance may be reduced, and rapid and efficient exchange of substrates is allowed [6]. Improved algal growth has been reported in the co-inoculation of bacterial strains (isolated from long-term laboratory algal cultures) than in algae alone [6], which suggests that the exchange of compounds between these microorganisms might improve their growth. Proximity, chemotaxis, adhesion, and mobility are important for the association microalgae-bacteria. Several reports have shown that aerobic heterotrophic organisms can use carbon compounds produced photosynthetically [7]. Photosynthesis, which is a reversible set of reactions, is inhibited by excessive dissolved oxygen; the oxygen consumption of bacteria decreases the tension in the microenvironment of algal cells, leading to more favorable conditions for algal growth [6]. Moreover, microalgae can use the carbon dioxide produced by bacterial mineralization for their in the photosynthesis.
To ensure the physical proximity between the microalgae and bacteria, both microorganisms need to be immobilized in a transparent matrix that must meet three requirements: (1) allow the entry of light needed by the microalgae to carry out their photosynthetic processes; (2) be small enough to allow diffusion of oxygen and nutrients; and (3) be large and heavy enough to prevent buoyancy and assure complete immersion in the growth medium [8]-[9].
Some reports of phenol bioremediation have evaluated concentrations between 250 and 500 ppm in the treatment system that were reduced to less than 5 ppm in some bacterial communities [10]. In the same way, Chlamydomonas reinhardtii and Chlorella vulgaris Beyerinck have demonstrated potential for utilizing phenol as a carbon source in an agar salt medium [11]. Another report introduced the ability of Stenotrophomonas species to degrade organophosphates and phenol simultaneously; they found a relation between the concentration of phenol and the optical density of the culture [12]. Likewise, a different study examined the biodegradation of phenol in a batch culture by pure strains of Paenibacillus sp; they screened several concentrations of phenol (from 100 ppm to 600 ppm) on a mineral salt agar medium and observed a 53.86% degradation of phenol at 500 ppm [13].
This work evaluates the phenol degradation capacity of microalgae-bacteria consortia using three bacterial strains isolated from the bay of Cartagena, Colombia, and the microalga Chlamydomonas reinhardtii. Thus, this work will contribute to biotechnology studies on bioremediation using Colombian native bacterial strains.
2. MATERIALS AND METHODS
2.1 Microorganisms and media
Strains were isolated in a mineral medium with phenol (ATCC, USA) as their only source of carbon and energy; they were also identified by 16 S rDNA gene, using the primers 1492r (5’-TACCTTG TTACGACTT) and 27F (5’-AGAGTTTGATCMTGGCTCAG) as previously described [9]. The company Genewiz (South Plainfield, NJ) conducted the sequencing of the PCR products. Isolates were identified as Stenotrophomonas spp. (C2), Microbacterium spp. (C7A), and Paenibacillus spp. (C7B), with 99% identity to Stenotrophomonas maltophilia strain KW 98 16S ribosomal RNA gene (Accession number JX262398.1), 99% identity to Microbacterium spp. BAB-4119 16S ribosomal RNA gene, (Accession number KJ778662.1), and 99% identity to Paenibacillus lactis strain NASCB-5 16S ribosomal RNA gene (Accession number KP100165.1), respectively.
A Tris-Acetate-Phosphate (TAP) medium (Culture Collection of Cryophilic Algae, 2014) was used for all the cultures: pure cultures and consortia as well as pure cultures and consortia with added phenol. Said medium contained a phosphate solution 1ml (K₂HPO₄ 28.8g 100ml-1, KH₂PO₄ 14.4g 100ml-1), salts 25ml (NH₄Cl 15g L-1, MgSO₄ 7H₂O 4g L-1, CaCl₂ 2H₂O 2g L-1), trace elements 1ml (H₃BO₃ 1.14g 100ml-1, ZnSO₄ 7H₂O 2.2g 100ml-1, MnCl₂ 4H₂O 0.5g 100ml-1, FeSO₄ 7H₂O 0.5g 100ml-1, CoCl₂ 6H₂O 0.16g 100ml-1, CuSO₄ 5H₂O 0.16g 100ml-1, (NH₄)6Mo7O₂₄ 4H₂O 0.11g 100ml-1, EDTA 5g 100ml-1 ), acetic acid 1ml, and tris 2.42g. All the reagents were obtained from a local supplier (Sigma-Aldrich, USA).
2.2 Growth monitoring and algae-bacteria ratio selection for the inoculum
The growth rate was evaluated in order to select the best conditions for degradation. Subsequently, individual and consortium growth curves of cultures (growing in a 15-mL TAP medium at 27°C and 100 RPM for 150 hours under continuous light) were obtained and compared to assess culture conditions. Optical density measurements were carried out at 620 nm for bacteria and 685 nm for microalgae in a UV-vis spectrophotometer (Agilent, CA, USA) to determine the growth rates for approximately 150 hours. The OD length used in the measurements was changed because of the maximum absorption wavelength of bacteria (620 nm) and microalgae (685 nm).
Moreover, microalgae cells were counted using a Neubauer chamber. Inoculum ratios of 1:1, 2:1 and 1:2 (for microalgae and bacteria, respectively) were evaluated. Here, 1 is equivalent to 100 µl and 2 equals 200 µl of inoculum. All cultures were monitored by light microscopy to analyze the interaction between the microalgae and the bacteria.
2.3 Phenol degradation evaluation
In order to evaluate the potential for phenol degradation of the consortia, an acclimation of the microorganisms to phenol at 50 ppm, 100 ppm and 150 ppm of phenol concentration was induced. TAP medium was added to all cultures containing different phenol concentrations. Their potential to degrade phenol was evaluated at 48 and 96 hours of incubation at 27°C with light by liquid chromatography (Agilent 1260 Infinity HPLC) using a ZORBAX Eclipse Plus C18 HPLC column under several conditions: injection volumes from 0.1 to 100 µl; column length up to 300 mm and internal diameter 0.05-8 mm; temperature range from 10°C below ambient to 80°C; maximum pressure 600 bar; and flow rate range from 0.05 to 5 mL/min at 600 bar and 5 to 10 mL/min at 200 bar. Multiple 1.5-ml samples of the pure and mixed cultures media were filtrated and then transferred to a vial for chromatography. The percentage of degradation was calculated by comparing the peak areas of the degraded and control samples.
3. RESULTS
Since the growth of Chlamydomonas reinhardtii is facilitated by saline media and adequate media formulation is important for consortia development, the performance of the microalgae was evaluated when growing in the presence of potentially halotolerant organisms and phenol degrading strains isolated from national ecosystems characterized by high salt concentrations, such as the bay of Cartagena in Colombia. Therefore, a TAP medium was selected for the analysis of the consortia.
First, the behavior of each bacterial strain and microalga was individually analyzed. In general, bacterial strains and Chlamydomonas reinhardtii displayed growth after 4 days of incubation at 30°C and after 6 days of incubation at 27°. The behavior of microalgae and bacterial strains was found to be similar as the lag phase for all strains was around 50 hours (Fig. 1), which may indicate that the selected medium is appropriate for consortia growth.
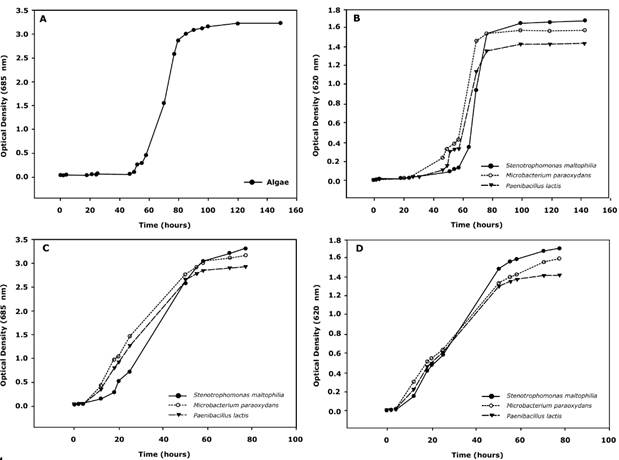
Source: Authors’ own work.
Fig. 1 Growth curves in a TAP medium. (A) Pure culture of microalgae Chlamydomonas reinhardtii, (B) Pure cultures of bacterial strains, (C) Microalgae Chlamydomonas reinhardtii-bacteria consortia in a ratio of 1:2 (bacteria-microalgae), (D) Microalgae Chlamydomonas reinhardtii-bacteria consortia in a ratio of 2:1 (bacteria-microalgae).
Moreover, the lag phase found in all the cultures is typical when phenol degradation is taking place, as the time for adaptation is remarkably longer than when different energy or carbon sources are available. This finding can improve the removal efficiency when the technology is scaled up, because a better strategy to acclimate each strain to phenol can be established. In addition, a reduction in the exponential phase of bacterial strains (compared to microalgae) was also found, possibly indicating a faster depletion and better adaptation of bacterial strains.
Light microscopy evaluations allowed to determine the presence of potential symbiosis in the consortia (Fig. 2 and Fig. 3).
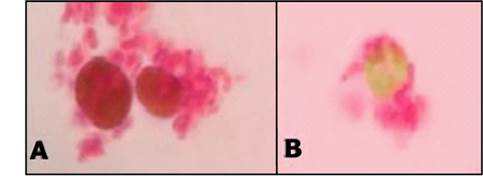
Source: Authors’ own work.
Fig. 2 Optical microscopy x100 of the consortium C7A-microalgae, fuchsine stained samples. (A) Pre-inoculum ratio, 2:1. (B) Pre-inoculum ratio, 1:1.
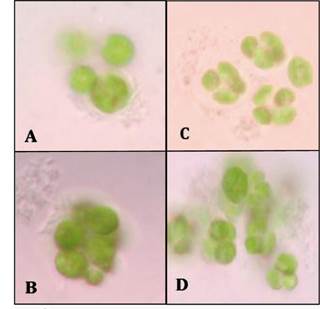
Source: Authors’ own work.
Fig. 3 Optical microscopy x100 of the consortia in a ratio of 2:1 (bacteria-microalgae). (A) Consortium C2-microalgae. (B) Consortium C7B-microaglae. (C) and (D) Consortium C7A-microalgae.
In all cases a contact between the microalgae and the bacteria was detected, possibly indicating the interaction between them. Nevertheless, the 2:1 ratio displayed greater and more cellular aggregations in the cultures as well as more contact between the species (Fig. 2). As a result, the growth curves of the 2:1 ratio were obtained and faster growth of the consortia, compared to individual cultures, was found. This behavior was reflected in a shorter lag phase (down to 10 h, an 80% reduction) and in half the time elapsed to reach the stationary phase (compared to individual cultures).
An initial visual comparison of the growth of different bacterial strains revealed interesting differences between them. It seems that the agglomeration of Microbacterium spp-microalgae is clearer than other strains. Moreover, the morphology of the algae does not seem to be affected by the bacterial strain present in the culture. The number of algal cells calculated in the 2:1 ratio culture (3.09*107 cells/mL) also displayed a significant difference compared with its counterpart in the pure microalgae culture (1.35*107 cells/mL in average). Growth curves were also obtained for each strain in TAP medium (Fig. 1). Our data suggested that the growth rates of the microalgae and the bacterial strains are independent of the consortia, possibly indicating that several factors such as metabolism associated with the species or growth rate do not significantly affect the evolution of symbiosis.
Phenol degradation was then evaluated at different phenol concentrations (50, 100, and 150 ppm) by measuring phenol at 48 and 96 h in different inoculum ratios for each bacterial strain (Fig. 4), in pure cultures as well as consortia. The initial concentration of phenol presents a significant effect on phenol removal, as 50 ppm of phenol results in the most significant removal by the Microbacterium spp-C. reinhardtii consortia; nevertheless, this effect seems to be mediated by the specific bacterial strain because phenol removal by Paenibacillus spp. is negatively affected by the presence of C. reinhardtii. Finally, our data shows that the phenol removal capacity of Stenotrophomonas spp. is facilitated by the presence of the microalgae, but it does not reach the level of Microbacterium spp-C. reinhardti.
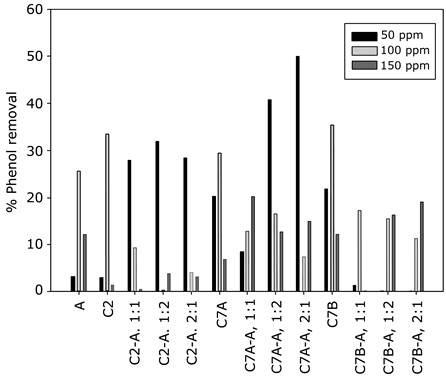
Source: Authors’ own work.
Fig. 4 Percentages of phenol removal in each culture at the evaluated concentrations. Abbreviations: A, microalga Chlamydomonas reinhardtii; C2, Stenotrophomonas maltophilia; C7A, Microbacterium paraoxydans; C7B, Paenibacillus lactis. 1:1, 1:2, and 2:1 denote the inoculum ratios of the consortia.
In general, it seems that high phenol concentrations negatively affect the synergy when microalgae are used; this was reflected in the fact that, in all the cases with 100 and 150 ppm, the phenol removal capacity of pure cultures is the same or higher than that of their consortia counterparts. Regarding the algae-bacteria ratio, our results confirm what was established without the presence of phenol, because the 2:1 bacteria-microalgae ratio displayed the most interesting removal capacity, especially the consortium Microbacterium spp-C. reinhardtii.
Phenol removal was finally evaluated using a modified TAP medium (without acetic acid and 100 ppm of phenol as the only source of carbon). None of the bacterial species displayed growth; nevertheless, the microalgae did grow in this medium and the culture coloration was less green than in a regular medium. After the first measurement, it was clear that the microalgae is the microorganism that leads the process of pollutant degradation.
4. DISCUSSION
First, our data suggest that the growth of the bacteria and microalgae under study was favored. A much faster growth was found in the consortia compared to pure cultures, which enables to claim that consortia enable growth stimulation. Furthermore, light microscopy revealed cell aggregations due to the tight contact between the microalgae and the different bacterial species, which may suggest symbiosis. Our results indicate that the formation of cell aggregations occurs as a result of the activation of defense mechanisms given the presence of other microorganisms in the same medium. Previous studies on associations of microalgae and bacteria have shown smaller cell aggregations in axenic cultures (only one species) than in their xenic counterparts (various species-consortia) and that cell formation (called floccules) takes place due to the growth stimulation that generates the interaction between different microorganisms [14].
Regarding the phenol degradation potential, the consortia created in this work present a better performance at low concentrations of the pollutant, which is one of the current bottlenecks in phenol remediation technologies [1]. The results of phenol degradation and microscopy enable to establish that a pre-inoculum ratio of 2:1 (bacteria-microalgae) is the most appropriate for the development of the consortia. Likewise, the most efficient consortium, in terms of phenol removal at 50 pmm, is developed with the strain Microbacterium spp, which achieved a removal percentage of approximately 50% (Fig. 4); this may indicate that 100 and 150 ppm concentrations induce resistance mechanisms instead of biodegradation metabolism.
The interactions between microalgae and bacteria are complex, including competition for resources and the production of extracellular polymeric substances and exudates [15]. Furthermore, bacteria tend to be effective competitors for resources because they have a rapid growth rate, a greater volume ratio per surface area and rapid rates of nutrient intake [15]. Some studies report competitive interactions where the inhibition of bacterial growth (Vibrio alginolyticus) is observed in alga cultures (Skletonema costatum) as a result of competitive exclusion [16]. Therefore, the dynamics of interaction between populations should be taken into account to create mixed cultures. The growth of the consortia observed in this work may indicate a symbiotic relationship between the species due to the cellular aggregations. Nevertheless, such aggregations may also be caused by resistance mechanisms activated by the microalgae.
5. CONCLUSIONS
Our results suggest that both growth rate and lag phase are different between microorganism growing alone or in consortium. Interestingly, the inoculum ratio 2:1 (bacteria-microalgae) was shown to be the most robust considering that both microorganisms improved their growth. In addition, our data revealed the highly potential of using bacteria-microalgae consortia for phenol degradation, where consortia displayed a better performance at low phenol concentrations. The data presented in this work may be relevant for the bioremediation of environments polluted with hydrocarbons, and it offers the advantage of using native Colombian bacterial strains.














