1. INTRODUCTION
In this study, we address the Relation Extraction (RE) problem as follows: For a given sentence S, the RE problem is a classification problem, where the goal is to predict a semantic relation r between e1 and e2, both entities in S, following previous research, mainly [1], as our baseline model. RE plays a key role in information extraction from unstructured text, and it has a wide range of applications in many domains [2], [3], [4].
The rapid growth of unstructured text data and the valuable knowledge recorded in them has generated considerable interest in automatic detection and extraction of semantic relations [5].
Although many studies have been conducted to develop supervised relation extraction models [6], [7], [8], [9], neural network-based approaches have been proposed for relation extraction, in particular with deep learning, e.g., Recursive Neural Networks [10]-[12], Recurrent Neural Networks (RNNs) [13], [14], [15], and Convolutional Neural Networks (CNNs) [1], [2], [16], [17], [18].
Deep Learning (DL) has demonstrated its efficiency in improving the RE task.
Specifically regarding relations in English language, the deep learning (DL) models have been trained with little or no domain knowledge, and several studies have implemented DL methods for relation extraction from texts. However, depending on the language and domain of deep learning models for relation extraction, the following challenges may arise: (a) a lack of training samples in some languages and domains and (b) the generalization of model in a domain with different types of relations.
Against this backdrop, some researchers have been successful in performing RE for a specific domain. They have utilized large amounts of labelled data. However, there are insufficient labelled data for certain domains and languages. Therefore, domain adaptation, domain shift, domain bias, and domain transfer are used to perform relation extraction an unseen target domain or language. However, the factors and conditions that are appropriate for training and testing DL models with different types of datasets in a target domain or language should be explored. Therefore, transferring well-trained DL models to other domains remains a challenge.
This paper presents a baseline DL model [1] and an experiment conducted using multiple representations from the biomedical domain. To address both challenges mentioned above, we describe the impact of some biomedical datasets, the generalization capability of the deep learning model, and compare its performance when some representations of the baseline model are modified for the biomedical domain.
This paper is organized as follows.
Section 2 presents a DL model based on [1], [16] for relation extraction, considering the potential of DL for RE tasks in bioinformatics research (biology, biomedicine, and healthcare). Section 3 describes a suitable way to adapt a well-trained model to the biomedical domain.
Section 4 details the experimental setup and the evaluation of the baseline model on multiple public PPI, DDI and CPI corpora. The last two sections discuss and summarize the representations of the impacts and the behavior of the baseline model on the datasets.
2. RELATED WORK
Traditionally, Relation Extraction (RE) has been a classification problem that occurs between two or more named entities in the same sentence that have a semantic relationship. Depending on the number of semantic relation classes, RE tasks can be binary or multi-class. In this study, we considered a binary relation extraction task in the biomedical domain. [19], [20], [21], [22].
Said task achieves a high performance with supervised approaches; however, it needs annotated data, which is time consuming and entails intensive human labor. Recently, models based on deep neural networks, such as CNNs and RNNs, have shown promising results for RE.
For example, in [1], the authors explored RE without exhaustive pre-processing. They employed a CNN and observed that any automatically learned features yielded promising results and could potentially replace the manually designed features.
In turn, in [10], [23] other authors proposed a DL approach with a RNN architecture and a matrix-vector word representation to explore the impact of the lack of explicit knowledge about the type of relation. Likewise, a study [11] compared the capabilities of CNN and RNN for the relation classification task. We reviewed other articles on relation extraction without specific domain. We also surveyed some models in the literature classified by network architecture and dataset. We hope this survey provides an overview to select a baseline model.
In Table 1, the baseline is marked in boldface, the CNN and RNN-based Models are learned on SemEval 2010, and the category Others is relevant for Deep Learning and RE tasks.
Table 1 Survey of studies into RE tasks using Deep Learning approaches

Source: Created by the authors.
Most of the studies we reviewed are concerned with English language, and their models have not been extended to other languages or domains. There is a variety of possible relations between domains and languages, characterized by their own syntactic and lexical properties.
Nevertheless, the notion of a relation, what it “means”, is inherently ambiguous [35]. Many efforts have been devoted to biomedical relation extraction, whose goal is to discover valuable knowledge about proteins, drugs, diseases, genes, adverse effects, and other biological interactions from unstructured free text [36].
3. METHODOLOGY
In this section, we introduce a model architecture based on Convolutional Neural Networks (CNN) and modifications that allow other representations.
3.1 Model
Based on the literature review above, with better performance architecture for RE tasks is CNN [37]. The model used in this paper contains three components: (a) a convolutional layer with multiple windows sizes, (b) a max pooling layer, and (c) a fully connected layer with dropout and softmax. Additionally, the input data undergoes a pre-transformation to vector representation. The transformation from words to vector representations has been described by several authors [38], [39].
We considered the model in [1] as baseline model, and trained it to automatically find relevant information (features or patterns) in a source sentence to predict semantic relations. We also added several modifications to keep some key elements of the baseline model that are consistent and consequent with a RE task. The diagram of the baseline model is shown in Fig. 1.
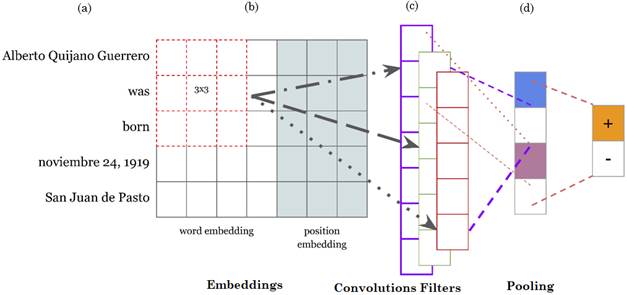
Source: Created by the authors.
Fig. 1 Diagram of the CNN-based model architecture implemented in this case study. (a) Words from a sample sentence represented as vectors (Input layer). (b) Vector representation where convolutional kernels are applied (Convolutional layer). (c) Vector pooling resulting from convolutional kernel. (d) Max values representative of the relation.
CNN-based architectures learn semantic information from sentences in the hidden layers during training. Although the extraction of semantic information is not previously known, the convolution layers learn features in the representations of the source domain. In the baseline model, let S be an input sentence that could be represented as S = {w1, w2, w3…wn}, where wi is the ith word in S; and let V be the vocabulary size of each dataset and Vxd, the embedding matrix with a d dimensional vector from pre-trained word embeddings.
Since we aim to compare the modifications and addition of representations to the baseline model in the biomedical domain, we used different kernel sizes and softmax function modifications; afterward, we included multi representations that captured different characteristics from the input. All of them were gradually changed. Then, for each wi in S, the distributional and not distributional representation was obtained and concatenated in a vector. As result, a matrix representing S was processed by the model in order to perform the classification.
This is a crucial part of our study in order to extract semantic relations, we needed to change the preprocessing, context length, paddings, kernel sizes, and validate implementation of baseline model.
Moreover, each word vector was supported by its corresponding information.
3.2 Representations
Representations have been effective tools to address the growing interest in DL for NLP tasks. While classical techniques used feature engineering and exploration to provide a more qualitative assessment and analysis of results from the point of view of computational linguistics, DL models learn features automatically.
In this paper, before S is processed by the baseline model, it is transformed in the form of a vector to capture different characteristics of the token; nevertheless, more information could be obtained from sentences to enhance automatic characterization via CNNs.
Multi-representation in DL models must be robust, and they should perform a satisfactory relation extraction in similar tasks across different domains. We used the following representations to add characteristic elements of sentences.
Word Embedding: It is employed to capture syntactic and semantic meanings of words in distributed representations.
In characterized by their own syntactic and lexical sentence S, every word wi is represented by a real-valued vector.
These word representations are encoded in an embedding matrix Xd, where V is a fixed-sized vocabulary.
Unfortunately, said word representations usually take a long time to train, and freely available trained word embeddings are commonly implemented [40]. We used pre-trained word2vec [38], Glove [39], and FastText [41] to conduct the experiments.
Position Embedding: In RE tasks, the words close to entities are usually informative and determine the relation between entities. We prove the relative position of words an entities similar to [1].
We used the relative position of both entity pairs. Apparently, it is not possible to capture such structural information only through semantic and syntactic word features. It is necessary to specify which input tokens are the target nouns in the sentence and where they are placed.
The position characterized by their own syntactic and lexical of entities is a relative distance, which is also mapped to vector representations.
4. EXPERIMENTS
These experiments are intended to show that DL models (baseline) for Relation Extraction (RE) can be adapted to another domain using multi-representation. First, we introduce the datasets and the metrics to evaluate precision: recall and f1-score. Next, we describe the parameters of the baseline model, the evaluation of the multi-representation, its effects, and performance on the data. Finally, we compare the performance of the baseline model with the modified model.
4.1. Datasets in the biomedical domain
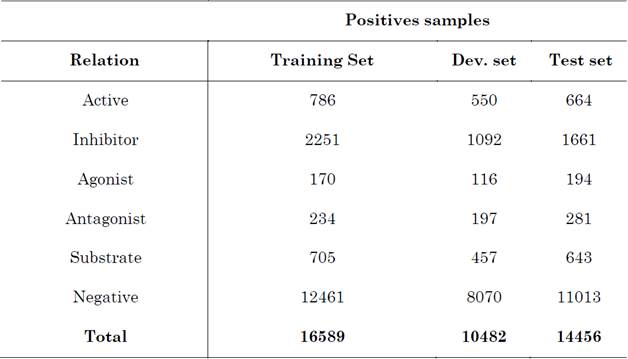
In this study, we explored RE tasks focused on the biomedical domain, especially relations such as protein-protein interactions (PPIs), drug-drug interactions (DDIs), and chemical-protein interactions (CPIs). Several scarce resources were utilized to adapt a pre-trained model.
We used three annotated corpora in the biomedical domain. All of them are publicly available and detailed below.
To extract semantic relations regarding Adverse Drug Effects, a subtask of DDI was applied to the corpus ADE-EX, as follows. The sentence “we report two cases of pseudoporphyria caused by naproxen and oxaprozin” contains a semantic relation of the type Adverse Drug Effect between pseudoporphyria and oxaprozin.
In turn, in BioInfer, we used the Protein-Protein interaction task to find semantic relations in the sentence “snf11 a new component of the yeast snf-swi complex that interacts with a conserved region of snf2”, where snf11 and snf2 are two named entities that represent proteins.
Likewise, in the corpus ChemProt, Chemical-Protein interactions are annotated. For example, in the sentence “Discovery of novel 2-hydroxydiarylamide derivatives as TMPRSS4 inhibitors”, 2-hydroxydiarylamide is a chemical and TMPRSS4 is a protein with a semantic relation to said chemical. For protein-protein interactions (relations), we used the BioInfer dataset [42]. For adverse drug events, we used the ADE corpus [43]; and, for chemical-protein interactions, we used the ChemProt corpus [44]. The detailed information of each dataset is listed in Table 2, Table 3 and Table 4, respectively.
BioInfer, a public resource providing an annotated corpus of biomedical English, is aimed at the development of Information Extraction (IE) systems and their components in the biomedical domain.
The ADE (Adverse Drug Effect) corpus consists of MEDLINE case reports annotated with drugs and conditions (e.g., diseases, signs and symptoms), along with untyped relationships between them.
ChemProt consists of PubMed abstracts annotated with chemical and protein entities. The relations were annotated with 10 chemical-protein relations. According to the shared task description, only 5 out of 10 semantic relation types would be evaluated.
Other important datasets for our study are SemEval-2010_Task_8 datasets [45] and ACE 2005 from LDC (Linguistic Data Consortium) [46]. Both were used in the baseline model, and their statistics are presented below.
SemEval 2010 task 8 is focused on multi-way classification between pairs of nominals. The task was designed to compare different approaches to semantic relation classification.
ACE-2005 consists of 6 main sources: broadcast news (bn), newswire (nw), broadcast conversation (bc), telephone conversation (cts), weblogs (wl), and usenet (un).
reACE, (Edinburgh Regularized Automatic Content Extraction) consists of English broadcast news and newswires with several annotated entities, such as organization, person, fvw (facility, vehicle or weapon), and gpl (geographical, political or location), along with relationships between them. Relationships are classified into five types: general-affiliation, organization-affiliation, part-whole, personal-social, and agent-artifact.
4.2 Measures
For the relation classification task, we used the F1-score as our measure for evaluation. The F1-score is defined as the harmonic mean between precision (P) and recall (R), such that, Precision = TP/ (TP+FP). Precision is the ratio of correctly predicted positive relations to the total predicted positive relations. In turn, Recall is the ratio TP / (TP + FN). Recall is the intuitive ability of the classifier to find all the positive samples. The F1-score is the weighted average of Precision and Recall.
F1 Score = 2*(Recall * Precision) / (Recall + Precision), where TP, FP and FN are true positives, false positives, and false negatives, respectively.
4.3 Hyperparameters and resources
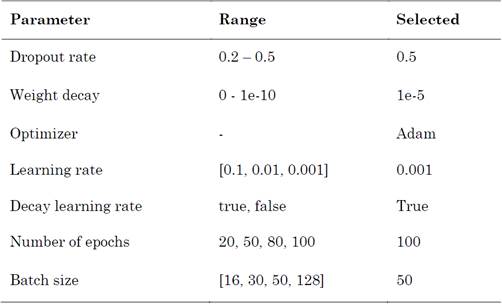
We considered a baseline model as a traditional approach to word representations and a CNN model with several windows without the combination of multi-representation. The benefit of multiple window sizes has been demonstrated; here, we used {3, 4} and {2, 3, 4, 5} to generate features. We tested several word representations with sizes d=50 and d=100, while the dimensionality of entity position indicators was d=20. Other parameters are listed in Table 5.
5. RESULTS AND DISCUSSION
The performance of the datasets SemEval 2010, ADE, BioInfer, Chemprot, and reACE is presented in Table 6 with the F1-score. Overall, we can make two observations: (1) The baseline model is not stable for each dataset. Although the predictions of the baseline model are not sufficient to establish what happened, the performance is meant to provide a neutral benchmark to measure the effects of adaptation changes. (2) Our models did not produce, in terms of performance, results comparable to those reported with the baseline model when biomedical domain datasets were used. ADE and reACE contain an imbalanced class distribution, which is perhaps the reason behind their 99.56 and 25.24 performance, respectively.
Table 6 Weighted F1-scores of the baseline and the four dataset variations (baseline score marked in boldface)
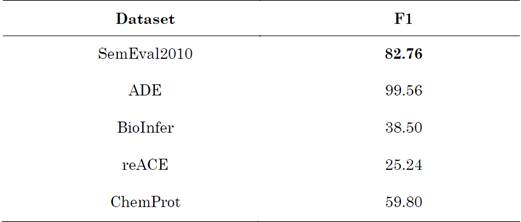
Source: Created by the authors.
Our hypothesis is that, as sample relations are extracted from an unbalanced corpus, our baseline model is more sensitive and a significant performance gap is produced. Our model, on the ADE corpus, achieved a high F1-score; however, the variation between the lowest and highest values of F1 in other datasets does not guarantee a superior performance.
This is perhaps not surprising since in-domain datasets contain short fragments of texts with scarce grammatical information, from which convolutions can capture relevant biomedical information and achieve a high F1. Moreover, after comparing CNN performance, we hypothesize that general and biomedical domain have equally or similarly difficult for RE task, when there is a difference across domain and domain models.
With supervised domain training and the model needs to capture knowledge and learn automatically features from the target domain. While we suspect there is still room for improvement, without utilizing domain specific information, the datasets may differ in ways we cannot account for with our reasoning.
Our baseline model exhibits two main aspects: First, unbalanced corpora have a negative impact on the F1-score.
More importantly, using a corpus with a balanced proportion of positive to negative relations can result in a better performance. Second, there are a number element in a deep learning model implementation, which makes exact replication of the results difficult, particularly performance results, but we compared our modified model from CNN base model (baseline), and the performance was quite similar. We believe our performance can be attributed to (1) vector representation and (2) class imbalance from the dataset. Thus, in-domain word embeddings and position embedding combinations are better for our model than out-of-domain word embeddings, although they cannot achieve results comparable to those of the baseline.
We presented a multi-step reasoning to train a model for other domains in cases in which data with other distribution and classes is available and the task is the same.
We also showed that our reasoning for model adaptation did not achieve a performance similar to that of the baseline model. Therefore, we carried out corpus-based exploration to address the adaptation of a deep learning model. We evaluated the DL model on different datasets. We also tested the deep learning model to extract semantic relations between entities implementing a similar experimental setup to that in the study by Zeng et. al [1].
After training and testing, our DL model should have learned how to extract semantic relationships due to the automatic learning of similar in-domain and out-of-domain features. However, our results confirm the need for a balanced dataset and additional information about.
the in-domain task. The proportion of positive and negative relations and the number of annotated data in the samples are different in each dataset (ADE, BioInfer, reACE, Chemprot, and SemEval).
Nevertheless, the problem of class imbalance between datasets has been reported in the literature [47].
We observed that the model is sensitive to word representations, which plays a significant role in model training. There are several embedding representations: position embedding (which represents the relative positions of entities and words in the sentence), medical and biological embedding (which contains specific information) in-domain word embedding (which includes methods that can generate domain-sensitive word embeddings).
Future studies can consider a similar reasoning, exploring, with combinations, different word representations (static, contextualized, with domain knowledge, and others).
6. CONCLUSIONS
In this paper, we proposed a DL model adapted to a new domain, more specifically, RE task for biomedical domain. We used an architecture to transfer the RE task from the generic domain to a biomedical one. After pre-processing the dataset, we obtained experimental results on several benchmark datasets. Nevertheless, we cannot confirm any advantage of the proposed model because it did not achieve a similar performance on different biomedical datasets or results comparable to those of SemEval 2010, which reached an F1-score of 82.76 (Baseline). Even though reACE, BioInfer, Chemprot, and ADE exhibited F1-scores of 25.24, 38.50, 59.80, and 99.56, respectively, these outputs cannot be rejected. We also analyzed the error and discuss the reasons behind our results.
Finally, our study explored different representations and results to avoid the duplicity of research efforts in the development of future systems.