Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Infectio
Print version ISSN 0123-9392
Infect. vol.15 no.3 Bogotá July/Sept. 2011
1GEPAMOL, Centro de Investigaciones Biomédicas, Universidad del Quindío, Armenia, Colombia
2Maestría en Microbiología, Universidad de Cartagena, Cartagena de Indias, Colombia
3Secretaría de Salud, Armenia, ColombiaRecibido: 08/02/2011; Aceptado: 03/08/2011
Abstract
Introduction: PCR detection offers a good opportunity to obtain fast results which is a priority in tuberculosis control programs.
Objectives: We assayed an in-house PCR method based on the detection of mycobaterial IS6110 gene in clinical samples of patients with pulmonary and extrapulmonary tuberculosis to demonstrate its usefulness and reliability in the setting of a middle-resource region with high tuberculosis prevalence.
Materials and methods: Pulmonary (n=317) and extrapulmonary (n=41) samples were collected from 358 patients with clinical suspicion of tuberculosis. All samples were processed to detect acid-fast bacilli by microscopy, culture on solid media and PCR. To remove PCR inhibitors, three washing steps of the decontaminated pellet were included before mycobacterial cell lysis.
Results: The overall sensitivity was 96% in clinical samples, and specificity was 100% for our in-house method in pulmonary and extrapulmonary samples. No inhibition was found among samples that were PCR negative, but culture positive for Mycobacterium tuberculosis. No false positives were found.
Conclusions: In-house PCR in a middle-income setting region, with simple and strictly controlled methods, could efficiently complement conventional bacteriological tools for the rapid diagnosis of tuberculosis, especially in paucibacillary and extrapulmonary samples.
Key words: Tuberculosis, Polymerase Chain Reaction, pulmonary tuberculosis, Mycobacterium tuberculosis.
Resumen
Introducción. La prueba de PCR ofrece la posibilidad de obtener resultados rápidos, una prioridad para los programas de control de la tuberculosis.
Objetivos.Evaluar el método de PCR basado en la detección del gen IS6110 de las micobacterias en muestras clínicas de pacientes con tuberculosis pulmonar y extrapulmonar, para demostrar su utilidad y confiabilidad en un sitio de desarrollo medio con alta prevalencia de tuberculosis.
Materiales y métodos. Se recolectaron 317 muestras pulmonares y 41 de origen extrapulmonar, de 358 pacientes con sospecha clínica de tuberculosis. Todas las muestras se procesaron para detectar bacilos ácido-alcohol resistentes por microscopía, cultivo en medio sólido y PCR. Para eliminar los inhibidores de la PCR se lavaron tres veces antes de realizar la lisis de la pared de la micobacteria.
Resultados. La sensibilidad total fue de 96% y la especificidad de 100% para este método "casero" en muestras pulmonares y extrapulmonares. No se encontró inhibición entre las muestras que fueron negativas por PCR pero con cultivo positivo para Mycobacterium tuberculosis. No se hallaron falsos positivos.
Conclusiones. La implementación de la PCR "casera" en regiones de desarrollo intermedio como método simple y estrictamente controlado, podría complementar eficientemente las herramientas bacteriológicas tradicionales para el diagnóstico rápido de la tuberculosis, especialmente en muestras paucibacilares y de origen extrapulmonar.
Palabra clave: tuberculosis, reacción en cadena de la polimerasa, tuberculosis pulmonar, Mycobacterium tuberculosis
Introduction
Tuberculosis is still a serious public health problem in many countries even with the existence of preventive measures to avoid its increase in the community. In Colombia, 11,047 new cases of tuberculosis (24.2 cases per 100,000 population) were reported (1). In Armenia (296,000 inhabitants), a city located at the central western part of Colombia, reported an incidence of 62.6 new cases per 100,000 population during 2009, compared with the national rate of 24.26 per 100,000; consequently, this geographic area has been classified as a high prevalence area for tuberculosis (2).
Additionally, multidrug-resistant strains increased notoriously in 2006-2008 in many cities of the country, including Armenia, with 0.5-2.2 %(3). There is a great concern of public health authorities about the dissemination of these strains, and they have urged the need for early diagnosis of active cases.
Rapid diagnosis of mycobaterial infections is currently based on slide examination of acid fast bacilli (4). This procedure is not very sensitive, especially in paucibacillary clinical samples and does not discriminate between Mycobacterium tuberculosis and other Mycobacterium species, like M. avium, a frequent cause of mycobacterial infection in immunosupressed patients (5).
Mycobacteria culture remains the gold standard for diagnosis, but it is expensive and demands more than 8 weeks to obtain a definitive identification. There are concerns about the availability of commercial PCR methods in poor and middle-income countries because in-house PCR assays produce highly variable estimates of sensitivity and specificity (6). A previous meta-analysis of various protocols for PCR-based diagnosis of pulmonary tuberculosis identified a few factors associated with improved diagnostic accuracy, and others that did not make a substantial difference and recommended that future development of PCR tests, to detect M. tuberculosis from sputum specimens, should take into consideration these test characteristics as a way of improving the accuracy of in-house tests to diagnose pulmonary tuberculosis (7, 8).
Therefore, we standardized and evaluated an in-house simple PCR method for the detection of the IS6110 gen specific for Mycobacterium tuberculosis complex and demonstrated that it is a feasible, rapid and economic method that can be applied in local conditions in a regional laboratory in highly endemic areas.
Material and methods
Ethical aspects, patients, and clinical specimens: Patients were informed about the study, and written informed consents were obtained for each one of them or their next of kin, according to the regulation number 008430 of 1993 of the Ministry of Health of Colombia.
Clinical samples were collected prospectively and consecutively from all patients with clinical suspicion of tuberculosis from January to December 2007. Patients were admitted to the University Hospital "San Juan de Dios" in the city of Armenia. Patients included in this study were not likely to be HIV positive.
From the 358 clinical specimens, 227 were sputum, 90 were bronchoalveolar lavage fluid, 4 were blood, 12 were gastric fluid aspirate, 15 were urine, 3 were cerebrospinal fluid, 3 were ascetic fluid, and 4 were abscess secretions. All samples, except blood, were stained by the Ziehl- Nielsen smear technique and examined for the presence of acid fast bacilli (AFB) according to standardized methods (9). All clinical specimens were processed in the Centro de Investigaciones Biomédicas, Universidad del Quindío.
Clinical diagnosis of tuberculosis was established by patient histories, clinical and radiological findings, and response to antituberculosis drug therapy. Response to antituberculosis treatment was recorded according to national guidelines (10). Only good quality sputum samples belonging to HIV-negative patients were studied. True positive samples were positive for M. tuberculosis in culture with or without AFB smear positive or were AFB positive with a negative culture, but patients showed a good clinical and radiographical response to antituberculosis treatment. All samples with negative cultures for mycobacteria or with positive AFB, but with no response to antituberculosis treatment were considered true negatives for M. tuberculosis.
Specimen processing: Sputum and urine samples were diluted v/v in 10% sodium phosphate buffer, incubated during 2 hours, and then homogenized in vortex and concentrated by centrifugation (4,000g for 30 minutes). Bronchoalveolar lavage fluid samples were treated with 10% sodium phosphate v/v, incubated during 2 hours and then homogenized in vortex and concentrated by centrifugation (1,500g, 10 minutes). Supernatant was recovered and centrifuged (4,000g, 30 minutes). Pellet was suspended in water again and divided in two aliquots for DNA extraction and culture. Blood samples were centrifuged (1,500g, 10 minutes), then the red blood cells were discarded and 0.4% sodium deoxicholate was added and the mixture was incubated during 30 minutes at room temperature. After this, the mixture was divided in aliquots for PCR and culture. Pellet was suspended again in 400 µl of TE buffer 1x (10 mM Tris-HCl pH 8,0 / EDTA 1,0 mM) and the tube was placed in a boiling water bath for 20 minutes according to national guidelines for tuberculosis laboratory techniques (11).
DNA extraction: 400 µl of sample suspended for a second time in TE buffer were added to 50 µl of lysozyme 10 mg/ml and incubated for 2 hours at 37°C. Then, 10 µl of proteinase K (10 mg/ml) were added and incubated for 10 minutes at 65°C to inactivate DNase and RNases. After that, 100 µl SDS 10% - CTAB 4% NaCl 5 M were added to dissolve the polysaccharides of cell walls of mycobacteria and incubated for 10 minutes at 65°C, and 750 µl of chloroformisoamil (24:1) were added and then centrifuged 12,000g for 15 minutes at room temperature. Supernatant was recovered and 500 µl of isopropanol were added and incubated for 1 hour at -20°C and then centrifuged for 15 minutes 12,000g at 4°C. Finally, the DNA was precipitated with 1 ml ethanol, washed twice and suspended again in 25 ml TE buffer (10 mM TRIS-HCl pH 8,0 / 1,0 mM EDTA). One sample without template was processed in 400 µl of TE as a control for contamination during the extraction step.
Mycobacterium tuberculosis culture and identification: M. tuberculosis cultures were derived from clinical samples. Samples were treated with NaOH and inoculated by duplicate in Lowenstein- Jensen and Ogawa-Kudoh medium (LJ-OK) using standard procedures. Growth assessment was performed at two, four, six and eight weeks. Identification of mycobacteria species was carried out by using conventional phenotypic tests (12).
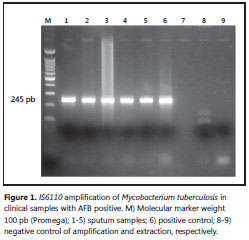
PCR procedure: A reaction mixture of 50 µl containing 5 µl of primers (0.5 µmol each INS-1 1 5’–CGTGAGGGCATCGAGGTGGC-3’ and INS-2 5’-GCGTAGGCGTCGGTGACAAA-3’), 5 µlTaq polymerase buffer 10x (25 mM Tris HCl [pH 9,5], 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, bovine serum albumin [1 mg/ml], 4 µl of dNTPs 2.5mM, 3 µl of MgCl2 25mM, 0.25 µl of Tucan Taq DNApol and 2 µl of DNA and 25.7 µl of water). The DNA was denatured for 5 minutes at 94°C followed by 40 amplification cycles with an automated thermal cycler. Each cycle consisted of denaturation at 94°C for one minute, annealing of primers at 68°C for one minute, and primer extension at 72°C for two minutes. Presence of the 245 bp amplification product was analyzed by electrophoresis of 5 µl of the amplified mixture on a 1.5% agarose gel. The DNA was stained by ethidium bromide and photographed on a UV transilluminator (13, 14). Amplification protocol detected DNA amounts of tuberculous mycobacteria higher than 50 fg (theoretically about 10 bacilli). A 245 bp strong band at different concentrations of genomic M. tuberculosis DNA was observed.
Evaluation of inhibition of PCR: All available specimens that were positive for M. tuberculosis growth but negative by PCR, and a group of randomly selected AFB culture positive and negative specimens, were analyzed for inhibition of PCR. This was briefly achieved by performing a second PCR on these lysates, under the conditions described above, but with the addition of known template and primers. The detection limit of PCR was determined by preparing 10- fold serial dilutions of M. tuberculosis DNA containing between 50 ng and 5 fg. Ten microliters from each dilution were used for amplification as described above. A duplicate of each DNA extract was spiked with approximately 0.5 pg of M. tuberculosis DNA from the H37Rv reference strain as recommended by Kunakorn (15).
Statistical methods and criteria for diagnosis evaluation: The chi square test was used to analyze results. Confidence intervals were calculated for 95%. Statistical analysis was carried out with the Epi-info software, version 3.5.1 (CDC, Atlanta). Sensitivity, specificity, positive predictive value, and negative predictive value were calculated according to applicable tables.
Cost estimates: A global evaluation was performed based on data from the account department at the Hospital San Juan de Dios, in Armenia based on reported mean costs of hospitalization for 2008.
Results
In total, 354 samples were analyzed by AFB and culture, and 4 blood samples were analyzed only by culture and PCR. In these samples, 62 were culture positive for M. tuberculosis and among them, 12 were AFB negative (Table 1).
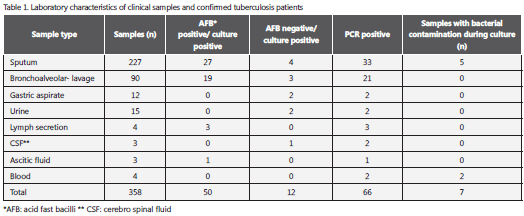
In seven patients, the culture was not performed due to a low sample amount or because results were non interpretable due to bacterial contamination, but with positive AFB smear and good response to antituberculosis treatment, they were also considered as true positive patients with tuberculosis (confirmed patients with tuberculosis). Consequently, the group of total true positive patients with tuberculosis was constituted by 62 patients M. tuberculosis culture positive and 7 patients with good response to specific antituberculosis therapy (total patients: 69, Table 2).
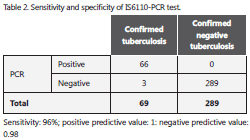
Amplification of 245 bp fragment of M. tuberculosis DNA was positive in 66 clinical samples between 358 tested (Figure 1). The comparison of the amplification with AFB and cultures are summarized in Table 1. Discrepant results in these samples were: two sputum samples and one bronchoalveolar lavage fluid that were PCR negative and culture positive, and two urine samples that were PCR positive, AFB negative and culture positive. Two blood samples were PCR positive, but no reliable culture result was obtained because there was bacterial contamination. PCR overall sensitivity was 96% (IC 95%: 90-100) and specificity was 100% (IC 95% 99-100) (Table 2).
A significant amount of cases with tuberculosis in the city of Armenia show a strong demand for third level medical care, which demonstrates the existence of a delayed diagnosis (16). As a consequence, patients showed a significant physical deterioration and a prolonged hospitalization period, which in turn increased the costs significantly. We estimated the hospitalization cost for the different types of tuberculosis as ranging from US$ 527 to US$ 10,883, with an average of US$ 4,623 per case; and hospitalization time varied between 4 and 54 days.
Discussion
Tuberculosis diagnosis is definitively established if the mycobaterial culture is positive, but this needs a long period to be able to obtain results (2 to 8 weeks). Additionally, 5-10% of cultures are negative, due to many factors as the low amount of bacilli, the non-viability of mycobacteria after decontamination procedure or the culture contamination with other bacteria (17). These conditions could explain the loss of 11% of our cultures. Moreover, the delayed diagnosis of tuberculosis delays treatment, aggravating the health condition of patients and raising the transmission risk. Although an AFB test positive is useful to detect paucibacillary cases, this assay is not sensitive enough as illustrated by 19% of our cases that were AFB smear test negative and culture positive. Other studies report similar results ranging between 20 and 50% of false negatives (7).
DNA amplification by PCR is a rapid and sensitive method for the detection of M. tuberculosis in clinical samples. One of the most important modifications that we ensured was to introduce a boiling step that simplifies the method. Previous studies have been retrospective in nature, focusing primarily on the determination of assay performance. We undertook a prospective study to examine the impact of molecular testing for tuberculosis in an effort to better determine the benefits and impact on patient care. PCR testing implementation on the initial two sputum samples collected during the first 24 hours of hospitalization would allow a faster diagnosis of tuberculosis. In this way, a specific clinical management could be rapidly implemented. In practice, smear-positive patients with a positive PCR indicate that the mycobacterial species is M. tuberculosis, and patients would be appropriately isolated and treated until their sputum samples became smear negative (18). Otherwise, a negative result would indicate the presence of a nontuberculous mycobacterial infection, and the patient could be released from isolation and treated more appropriately. Potential for positive impact on patient care with a reduction in the cost of care is apparent (19).
PCR may be useful in the diagnosis of extrapulmonary tuberculosis infections, particularly for meningitis. In our series, we found 17 cases with positive PCR, but AFB smear negative result. This is an important issue in the diagnosis of patients with clinical symptoms of tuberculosis. We found a 96% sensitivity, a very good result compared to previous papers of in-house PCR methods that reported sensitivities between 53 and 73% (20).
PCR inhibition, a frequent phenomenon in blood samples, was controlled in our method (21, 22). Bronchoalveolar fluid and urine samples did not show problems of specificity. The urine sample was from a patient with pulmonary tuberculosis and sputum AFB smear test was negative. Urine samples are easily obtained and they seem very useful to confirm tuberculosis before it is confirmed to be positive by culture.
Many cases of tuberculosis were diagnosed only by PCR. A blood sample was obtained in one patient with disseminated tuberculosis. In this case, the culture was contaminated with other organisms and diagnosis was reached only by PCR. Also, one of the urine samples came from a patient with pulmonary form and acid fast bacilli smear test negative. In another case, the urine sample came from a patient with acid fast bacilli smear test positive. PCR value on blood samples, and urine in pulmonary and extrapulmonary forms of tuberculosis, can be significant when it is difficult to obtain good quality samples (4, 5, 6, 7).
There is some debate about the use of IS6110 as a target for M. tuberculosis PCR because, in some regions, strains lacking this element can be up to 20% (23). In Colombia, one study found 7% of strains without this element (24). However, a previous study in the Quindío region reported the presence of this element in 27 M. tuberculosis strains (25).
Many variations on the use of PCR diagnosis have been described, some for commercial methods and others for in-house methods. Commercial methods tend to be advantageous because results can be obtained within 4-6 hours. However, costs for these commercial methods are expensive and require special equipment. Our in-house method requires two days and multiple steps in the process of sample preparation, such as extraction and DNA purification. However, this represents an advantage over commercial methods that use lysis buffers that are not very effective and have high levels of inhibition for Taq polymerase enzyme. The protocol used in our in-house method, including incubation with lysozyme during a long time (12 hours) and DNA purification with chloroform, isoamyl alcohol and ethanol, ensures a greater lysis of mycobacteria and less inhibition. Thus, this protocol obtains a highly pure DNA of good quality (not degraded by nucleases) that is enough for amplification and without inhibitors.
One critical aspect of in-house methods is the control of the contamination of the PCR reaction. We included negative controls for extraction and amplification steps to avoid false positives due to exogenous contamination. With these precautions, we suggest that the test is applicable in our laboratory for the identification of M. tuberculosis in clinical samples to confirm AFB smear-positive results.
In summary, nucleic acid amplification assays can be used on smear positive sputum samples and potentially on smear negative samples, if the clinical presentation is highly suggestive of tuberculosis. The most important application of our inhouse PCR that targeted IS6110 specific genomic elements of M. tuberculosis is a rapid diagnosis confirmation of clinical samples from pulmonary and extrapulmonary samples of cases with clinical suspicion of tuberculosis. In extrapulmonary samples, its sensitivity is the major advantage of PCR, and we showed that it can detect amounts of mycobacteria as low as 10 bacilli.
Acknowledgements
The authors wish to acknowledge the medical staff from the "Hospital Universitario San Juan de Dios" and Luz Piedad Quebrada for her technical support. The authors declare no conflict of interest with the manufacturers of reagents used in this study.
Correspondencia: Jorge Gómez-Marín, GEPAMOL, Centro de Investigaciones Biomédicas, Universidad del Quindío, Avenida Bolívar 12N, Armenia, Colombia. Dirección electrónica: gepamol2@uniquindio.edu.co
References
1. WHO. Global tuberculosis control: WHO report 2010. Geneva: World Health Organization; 2010. p. 204. [ Links ]
2. Secretaría de Salud Municipal de Armenia. Situación de salud en Armenia, indicadores básicos, 2007-2008. Informe anual del Programa de Control de la Tuberculosis, 2010. Armenia (Colombia): Alcaldía de Armenia; 2008. p. 22. [ Links ]
3. Secretaría de Salud Municipal de Armenia, Boletín Epidemiológico Armenia 2008. Informe de Gestión de la Alcaldía de Armenia (Quindío). Armenia: Alcaldía de Armenia, 2009:19. [ Links ]
4. Liu, K., W. Su, R. Perng, Clinical utility of polymerase chain reaction for diagnosis of smear-negative pleural tuberculosis. Journal Chinese Medical Association, 2007;70:148. [ Links ]
5. Tzoanopoulos D, Mimidis K, Giaglis S, Ritis K, Kartalis G. The usefulness of PCR amplification of the IS6110 insertion element of M. tuberculosis complex in ascitic fluid of patients with peritoneal tuberculosis. Eur J Intern Med. 2003;14:367-71. [ Links ]
6. Blanie M, Pellegrin JL, Maugein J. Apport de la PCR dans le diagnostic des tuberculoses extrapulmonaires. Med Mal Infect. 2005;35:17-22. [ Links ]
7. Sarmiento OL, Weigle KA, Alexander J, Weber DJ, Miller WC. Assessment by meta-analysis of PCR for diagnosis of smear-negative pulmonary tuberculosis. J Clin Microbiol. 2003;41:3233. [ Links ]
8. Pai M, Ramsay A, O´Brien R. Evidence-based tuberculosis diagnosis. PLoS Med. 2008;7:e156. [ Links ]
9. Organización Panamericana de la Salud. Manual para el diagnóstico bacteriológico de la tuberculosis. Normas y guía técnica. Parte I. Baciloscopia. . Washington, D.C.: Panamerican Health Organization; 2008. p. 64. [ Links ]
10. Ministerio de la Protección Social. Guía de atención de la tuberculosis pulmonar y extrapulmonar. Guías de promoción de la salud y prevención de enfermedades en la salud pública. Bogotá: Ministerio de la Protección Social; 2007. p. 105. [ Links ]
11. Instituto Nacional de Salud. Tuberculosis. Técnicas y procedimientos de laboratorio. Bogotá: Instituto Nacional de Salud; 2001. p. 51. [ Links ]
12. Organización Panamericana de la Salud. Manual para el diagnóstico bacteriológico de la tuberculosis. Normas y guía técnica. Parte II. Cultivo. Washington, D.C.: Panamerican Health Organization; 2008. p. 107. [ Links ]
13. Eisenach KD, Sifford MD, Cave MD, Bates JH, Crawford JT. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Am Rev Respir Dise. 1991;144:1160. [ Links ]
14. Puerto G, Castro CM, Ribón W. Polymerase chain reaction (PCR): a contribution for diagnosis of extrapulmonary tuberculosis and micobacteriosis. Infectio. 2007;11:97-100. [ Links ]
15. Kunakorn M, Raksakai K, Pracharktam R, Sattaudom C. Overcoming the errors of in-house PCR used in the clinical laboratory for the diagnosis of extrapulmonary tuberculosis. Southeast Asian J Ttrop Med Public Health. 1999;30:1:84-90. [ Links ]
16. Arciniegas W, Orjuela DL. Tuberculosis extrapulmonar: revisión de 102 casos en el Hospital Universitario San Jorge de Pereira, 2000- 2004. Biomédica. 2006;26:1. [ Links ]
17. Burman WJ, Stone BL, Reves RR, Wilson ML, Yang Z, El-Hajj H, et al. The incidence of false-positive cultures for Mycobacterium tuberculosis. Am J Respir Critical Care Med. 1997;155:321. [ Links ]
18. Davies AP, Newport LE, Billington OJ, Gillespie SH. Length of time to laboratory diagnosis of Mycobacterium tuberculosis infection: comparison of in-house methods with reference laboratory results. J Infect. 1999;39:3:205-208. [ Links ]
19. Kaul KL. Molecular detection of Mycobacterium tuberculosis: impact on patient care. Clin Chem. 2001;47:1553. [ Links ]
20. Flores LL, Pai M, Colford JM Jr, Riley LW. In-house nucleic acid amplification tests for the detection of Mycobacterium tuberculosis in sputum specimens: meta-analysis and meta-regression. BMC Microbiol. 2005;5:55. [ Links ]
21. Gunisha P, Madhavan HN, Jayanthi U, Therese KL. Polymerase chain reaction using IS6110 primer to detect Mycobacterium tuberculosis in clinical samples. Indian J Pathol Microbiol. 2001;43:395. [ Links ]
22. Verettas D, Kazakos C, Tilkeridis C, Dermon A, Petrou H, Galanis V. Polymerase chain reaction for the detection of Mycobacterium tuberculosis in synovial fluid, tissue samples, bone marrow aspirate and peripheral blood. Acta Orthop Belg. 2003;69:396-9. [ Links ]
23. Balamurugan R, Venkataraman S, John KR, Ramakrishna BS. PCR amplification of the IS6110 insertion element of Mycobacterium tuberculosis in fecal samples from patients with intestinal tuberculosis. J Clin Microbiol, 2006;44:5:1884. [ Links ]
24. Gómez-Marin JE, Leon Franco CI, Inirida Guerrero M, Rigouts L, Portaels F. IS6110 fingerprinting of sensitive and resistant strains (1991-1992) of Mycobacterium tuberculosis in Colombia. Mem Inst Oswaldo Cruz. 2002;97:1005-8. [ Links ]
25. Gómez Marín JE, Rigouts L, Villegas Londoño LE, Portaels F. Restriction fragment length polymorphism (RFLP) analysis and tuberculosis epidemiology. Epidemiol Bull. 1995;29:226. [ Links ]














