Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Infectio
Print version ISSN 0123-9392
Infect. vol.16 no.1 Bogotá Jan./Mar. 2012
1Curso de Graduação em Medicina Veterinária, Curso de Graduação em Medicina, Centro Universitário Serra dos Órgãos (UNIFESO), Teresópolis, RJ, Brasil.
2Departamento de Medicina e Enfermagem, Universidade Federal de Viçosa (UFV), Viçosa, MG, Brasil.
3Departamento de Bioquímica e Biologia Molecular, Universidade Federal de Viçosa (UFV), Viçosa, MG, Brasil.
4Curso de Graduação em Medicina, Centro Universitário Serra dos Órgãos (UNIFESO), Teresópolis, RJ, Brasil.
5New York University School of Medicine (NYU), New York, USA Work performed in the Departamento de Medicina e Enfermagem and Departamento de Bioquímica e Biologia Molecular of the Universidade Federal de Viçosa (UFV) and the Curso de Graduação em Medicina of the Centro Universitário Serra dos Órgãos (UNIFESO). There is no conflict of interest.
Recibido: 13/06/2011; Aceptado: 08/02/2012
Abstract
Chagas disease, an illness caused by the protozoan Trypanosoma cruzi, is clinically and epidemiologically important in Latin America, and particularly in Brazil. This article presents the main biological characteristics of Trypanosoma cruzi, emphasizing ultrastructural, morphological, evolutionary, transcriptomic, and proteomic aspects. With this purpose a literature review was conducted, which allowed for the construction of different sections of the text. The efforts to expand the knowledge of this protist biology may bring positive implications for the understanding of the pathogenesis, control, and above all, treatment of patients affected by this disease.
Key words: Trypanosoma cruzi; biology; transcriptome; proteome.
Resumo
A moléstia de Chagas, enfermidade causada pelo protozoário Trypanosoma cruzi, apresenta grande relevância clínica e epidemiológica na America Latina, com destaque para o Brasil. Neste artigo serão apresentadas as principais características biológicas do Trypanosoma cruzi, enfatizando-se os aspectos ultra-estruturais, morfológicos, evolutivos, transcriptômicos e proteômicos. Para tanto, foi realizada revisão da literatura, a qual subsidiou a construção de diferentes seções do texto. As investidas para ampliar o conhecimento acerca da biologia do protista poderão trazer implicações positivas para a compreensão da patogênese, do controle e, sobretudo, do tratamento dos pacientes portadores da moléstia.
Descritores: Trypanosoma cruzi; biologia; transcriptoma; proteoma.
Introduction
In 1909 a new species of parasite was discovered by the Brazilian physician Carlos Justiniano Ribeiro Chagas. He named it Trypanosoma cruzi in homage to Oswaldo Gonçalves Cruz who pioneered the study of parasitic and infectious diseases in Brazil. Due to his discovery and commitment to deepen the knowledge about the disease then called American trypanosomiasis, the recently discovered infectious disease was renamed Chagas disease, in his honor (1). To this day his not having been awarded the Nobel Prize in medicine or physiology after such contributions to tropical medicine is still discussed (1-3).
A century after his discovery, it is estimated that 300,000 new cases of Chagas disease are detected and 23,000 deaths are recorded yearly on the American continent (4), where the disease stands out in the systemic approach to human health, given that it represents the fourth largest social impact among all prevalent infectious and parasitic diseases (5). There are more than 100,000 individuals chronically infected by T. cruzi living in the United States, as a result of the migration of large groups of Latin Americans. Other cases were attributed to hemotransfusion and solid organ transplants in the United States, Canada and some European countries, where donors were not routinely screened for the protozoa till recently (6).
The mechanisms involved in the pathogenesis of Chagas disease are not yet fully elucidated. A number of factors such as the persistence of the parasite in human tissues, autoimmune response and microvascular alterations are now largely researched with the purpose of understanding the sickness and developing control and treatment strategies (7).
Knowledge about the biology of T. cruzi has helped understanding the pathogenesis and the clinical manifestations of the disease (8-10), as well as proposing diagnostic methods and searching for new treatments for affected patients (11,12). In fact, the recent T. cruzi DNA sequence amplification through polymerase chain reaction (PCR) technique has become the most sensitive diagnostic method for the disease, and as such the method of choice for detection of the protozoan in blood and organs (13). Furthermore, the recognition that T. cruzi DNA is present in various tissues, may help defining the ideal drug regimen and its optimal duration for patients chronically ill, what has not yet been established (14). Studies involving proteomic analysis are still under development, but already show good prospects concerning various aspects of Chagas disease (15).
T. cruzi - the etiologic agent of Chagas disease or American trypanosomiasis - is a flagellate protozoan, phylum Sarcomastigophora, class Mastigophora, order Kinetoplastida and family Trypanosomatidae, genus Trypanosoma, subgenus Schizotrypanum (16). The fact that it is eliminated together with the excreta of its invertebrate host had it included among the stercoralia, while other species of its genus belong to the salivaria, such as Trypanosoma brucei.
Based on these considerations, this paper aims to discuss the main biological characteristics of T. cruzi, emphasizing its ultrastructural, morphological, and evolutionary aspects, as well as the more recent investigations involving the transcriptome and proteome of T. cruzi.
Methods
The text is based on a literature review with strategy defined search. The articles were searched in the Scientific Electronic Library Online (SciELO) and the U. S. National Library of Medicine (PubMed) using the following terms: Strategy 1 - Trypanosoma cruzi + ultrastructure Strategy 2 - Trypanosoma cruzi + morphology Strategy 3 - Trypanosoma cruzi + life cycle Strategy 4 - Trypanosoma cruzi + proteome Strategy 5 - Trypanosoma cruzi + transcriptome From the total number of citations obtained (Table 1), the papers selected focused on the biology of the protozoan, with emphasis on the etiopathogenesis, clinic, diagnosis, and therapeutics of Chagas disease, adding up to 110 articles which served as reference to this paper. The articles were discussed and the information organized in sections - (1) characterization of the protozoan, (2) ultrastructure, (3) morphological aspects, (4) life cycle, (5) T. cruzi transcriptome analysis, (6) T. cruzi proteome analysis, and (7) final considerations.
In addition to the articles, Internal Medicine and Infectious Diseases textbooks were also consulted as part of the bibliographic research.
Results and discussion
Characterization of the protozoan
The species Trypanosoma cruzi is a heterogeneous population composed of diverse strains circulating in nature, with genetic variability observed among them (17-19). One of the first proposals of T. cruzi biological characterization was made by Brener (1977) (20), who described Y and CL strains as polar strains, representatives of the substantial intraspecific variability of the protozoan. More recently, the main criteria used in the characterization of the various strains of T. cruzi include haplotypes, discrete typing units (DTU), ribosomal DNA, schizodemes, zymodemes, bio demes, sensitivity to chemotherapics, virulence and pathogenicity, clinical and pathological forms of the disease, species of vertebrate hosts infected and geographic origin (19, 21-31).
A new attempt was recently made to unify the classification of T. cruzi strains (32), with the recommendation for the characterization of six groups called TcI to TcVI (for details, see reference 32). This classification took into account previous studies of various markers for many isolates and known strains; however a testing model protocol for classifying new isolates and strains has not yet been standardized.
Regardless of the classification method employed, it is important not to lose sight of the larger goal of the proposed groupings, which is establishing correlations between the infecting strain and the immunology, pathology, clinic, therapeutics, and epidemiology of Chagas disease (27,33,34).
Trypanosoma cruzi ultrastructure
The T. cruzi plasma membrane (PM) is composed of a lipid bilayer, in which several proteins are "immersed". Its main components are (19,28): proteins - irregularly distributed due to their displacement through the lipid bilayer (capping and patching movements), as observed in the trypomastigote forms -, phospholipids - phosphatidylcholine (most prevalent), phosphatidylethanolamine (PE), and phosphatidylinositol (PI), and in smaller quantities sphingomyelin (SM), phosphatidylserine (PS), cardiolipin (CLP), phosphatidic acid (PA), phosphatidylinositol phosphate (PIP), and phosphatidylinositol bisphosphate (PIP2), the latter important in the transduction of T. cruzi signal - and glycocalyx (thin, covering the outermost surface, consisting of glycolipids, glycoproteins, and lipopeptides). On the outer surface a developed cell coat is observable, three times thicker in trypomastigotes than in epimastigotes. The presence of glygopeptide-associated sialic acid is also described, playing an important role in the T. cruzi / macrophage interaction (26). On the internal side of the membrane, many microtubules are observed, extending from one end of the flagellate to the other (35). The use of modern microscopy techniques, such as cryofracture, has shown details of the membranes in different regions of the protist and in different morphological forms, where specialized macro-, micro-, and nano-membrane domains have been observed. In particular, intramembrane density differences were noticed, possibly related to the unequal presence of integral membrane proteins specific to each stage of development. For instance, in epimastigote forms, the flagellar membrane was found to contain a much smaller number of inserted particles than the rest of the membrane covering the body of the protozoan.
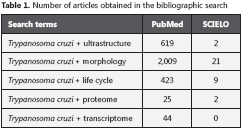
The T. cruzi cytoplasm presents different organelles (figure 1), including (36-38):
1. The single tubular mitochondrion, presenting irregular walls and cristae, its appearance depending on the evolutionary stage analyzed, having as limiting element a double mitocondrial membrane. The mitochondrion presents an intimate relationship with the kinetoplast - they are in fact considered as the same structure - with continuity between the walls of both organelles.
2. The kinetoplast, a structure peculiar to the order Kinetoplastida, contains high concentrations of the parasite´s extranuclear DNA, known as kinetoplastic or k-DNA. Unlike other orders of eukaryotes - which present roughly around 1% of cellular DNA in the mitochondria - T. cruzi can concentrate between 16 to 30% of its total DNA in the mitocondrial genome (39).
k-DNA is made up of approximately 5,000 to 20,000 minicircles and 50 copies of maxicircles per network, differing from nuclear DNA (nDNA) in base composition, molecular structure, and absence of histone bonds. kDNA network replication involves the release of the minicircles contained in this structure, enabling further replication and its reintegration to the network by the action of enzymes known as topoisomerases. The minicircles represent 95% of the molecules that make up kDNA, equivalent to 10,000 to 30,000 units according to the species and strains of trypanosomatid. Maxicircles are of lesser importance to the spatial maintenance of the kDNA (33), some mutants of the Trypanosoma genus not presenting them (40,41). Minicircles are heterogeneous in sequencing, despite their presence within the same species in relatively homogenous sizes (42,43). This heterogeneity within the kDNA networks is not absolute, there usually is a certain degree of homology among them. Analysis of the regions found in the minicircles reveals minirepeats, similar intraspecific sequences varying from 100 to 200 base pairs (bps) (44,45); the number of conserved regions is invariable for each species. Trypanosome cruzi possesses four copies of the conserved region, with 122 bp, arranged in compositions of repetitions positioned at 90º from each other, with three conserved sequence blocks (CSB) (44,46). The main function suggested for the maxicircles is encoding respiratory chain enzymes - such as ATPase and cytochromoxidase - and they are considered true representatives of mitocondrial DNA (43, 47), while the minicircles appear to code guide ribonucleic acids (gRNA) required for the editing of mitochondrial RNA (48,49), which in association with the minicircles has a function similar to that of the maxicircles in the coding of genetic information necessary for RNA editing.
3. The kinetosome, also called basal body, is the continuation of the flagellum, and is closely associated with the kinetoplast thanks to the connection between these two structures formed by numerous protein filaments.
4. The glycosomes - which according to Souza (2008) (50) - are spherical or elongated structures surrounded by a membrane, presenting a homogeneous membrane. Initially, these organelles were called microcorpuscles, in analogy to the catalase enzyme-containing structures found in mammalian cells that were later called peroxisomes. However, unlike the structure of mammalian cells, the main specific characteristic that gives name to this organelle in trypanosomatids is the presence of a majority of glycolysis enzymes. Glycosomes are currently considered a special type of peroxisomes in trypanosomatids, because in some species the presence of catalase was also demonstrated in this organelle.
5. The flagellum is made up of nine microtubule pairs arranged in a circle along with two central microtubules, all immersed in cytoplasmic matrix surrounded by a cellular membrane in its full extension, presenting, in the region where it is implanted in the cell body, an invagination called the flagellar pocket.
6. The Golgi structure is similar to mammalian Golgi complex and is engaged in protein glycosilation and membrane trafficking too (50).
7. The cytostome seems to be a specialized invagination of the plasma membrane that penetrates into the cell body and is involved in endocytic activity; especially in epimastigotes the cytostome is responsible for at about 85% of total endocytosis (50).
8. The flagellar pocket is a structure at the base of the flagellum and is responsible for endocytic and exocytic activities (51). In Trypanosoma brucei it is involved in many biological processes such as cell polarity, cell division, protein trafficking and immune evasion (52).
9. The reservosomes are described as a group of monolayers rounded or irregular structures containing several inclusions in their matrix. They are concentrated at the posterior region of the parasites and disappear during the transformation of epimastogotes into trypomastigotes. They have proteases and accumulate proteins from endocytosis (50).
10. The acidocalcisome is a vacuolar organelle able to transport protons and calcium. They contain calcium, phosphorus, sodium, potassium and zinc. They are involved with the storage of this elements and phosphorus derivative compounds as polyphosphate compounds. In addition they participate in pH homeostasis and osmoregulation (53).
The structure of the nucleus corresponds to the pattern observed in other eukaryotic cells (28,54), and may have varying shapes, depending on the parasite evolutionary form (19).
Morphological aspects
In order to adapt to the different interior microenvironments of its hosts, T. cruzi must undergo biological transformations, which cause structural and metabolic changes and enable the viability of the infection (55). The host animals of the T. cruzi cycle include a mammal and an invertebrate perforce hematophage which acts as a vector, belonging to the order Hemiptera, family Reduviidae, subfamily Triatominae (56). The protozoan has three evolutive forms (57-59), identified morphologically by optic microscopy, by the position of the kinetoplast in relation to the cell´s nucleus and by the emergence of the flagellum: amastigotes, epimastigotes, and trypomastigotes (36,60).
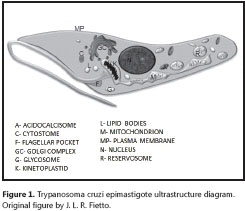
The amastigote (figure 2) is the intracellular form of T. cruzi, found in the tissues of the vertébrate host. It lacks an exterior flagellum and undulating membrane - on account of this, its movement is only by rotation - and it measures roughly 4.0 μm (between 2.0 and 6.5 μm) in diameter. It multiplies by longitudinal binary division every 12 hours, transforming into the blood trypomastigote approximately 11 to 13 hours prior to cell rupture. The amastigote forms, when released in the circulation, are also capable of infecting new cells (61). Additionally, it was recently demonstrated using genetically modified fluorescent protozoa, that the epimastigote forms are also infective (62,63). This same group of researchers demonstrated that although the epimastigote forms are as infective as trypomastigotes and amastigotes, there are three different forms of replication, differentiation, and release time (in relation to the conclusion of one cell cycle) (43,48).
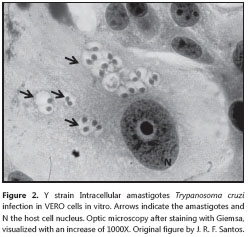
The epimastigote form is found in the digestive tract of the triatomine as well as in the anal glands of opossums. It presents a free flagellum, kinetoplast anterior to the nucleus and poorly developed undulating membrane measuring between 20 and 40 μm, including the free flagellum. The nucleolus is spherical, generally occupying a central position in the nucleus (54). It is quite mobile, presenting intense replicative activity - also by longitudinal binary division - and oftentimes forming rosacea. This evolutive form does not withstand the temperature of 37oC, with a better development in the range of 20-28oC.
The trypomastigote does not possess replicative capability, and corresponds to the extracelular infective form, located in both the invertebrate and vertebrate hosts as the metacyclic and blood trypomastigote, respectively. The metacyclic trypomastigote is found in the final portions of the intestine or in the Malpighi tubes of the insect vector. Morphologically, it resembles the thin forms of the blood trypomastigotes, measuring roughly 17 μm. It presents a large and central nucleus and a kinetoplast of high DNA density in a terminal posterior position. The flagellum is shorter - emerging from the basal corpuscle near the kinetoplast - and the undulating membrane is usually narrow. The blood trypomastigote is observed in the blood and other body fluids - such as the cerebrospinal fluid and lymph - of the vertebrate hosts. The flagellum emerges from the basal corpuscle near the kinetoplast, and is long (representing on average one third of the length of the protozoan.) The undulating membrane is narrow. It measures between 12 and 20 μm (including a free flagellum) (19,28,63).
Life cycle
The T. cruzi life cycle is complex (figure 3), and is marked by a series of transformations (28,36,64).
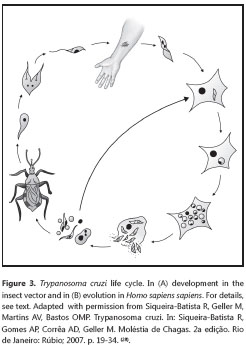
When feeding, the infected triatomine receives a significant amount of blood in its digestive system, that forces the elimination of the mass of accumulated excreta - consisting of feces and urine - which is normally deposited on the skin surface. These wastes contain the metacyclic trypomastigotes, which, by active movement and release of histolytic enzymes, actively penétrate the skin or mucosa. Should pruritus occur, there may be excoriation of the site - as a result of scratching - facilitating penetration of the protozoan; additionally, the parasite may be transferred to other regions of the skin or to the ocular mucosa. Once the metacyclic trypomastigotes reach the tissue of their vertébrate host, they are endocyted by the local mononuclear phagocytic system or by other, not typically phagocytic cell types, in a process known as induced phagocytosis. In the parasitophorous vacuole formed after penetration of the host cell, the trypomastigotes release proteic factors that act in the release of this organelle, using for instance, a cytotoxic protein (Tc-TOX) which presents some similarities to the perforins of the cytotoxic T lymphocytes (36), in addition to the trans-sialidases, very abundant enzymes in this protozoan. Although the trans-sialidases have been implicated in the penetration of the host´s cells, their function in this stage has not been entirely elucidated. On the other hand, there is evidence that trans-sialidase-induced removal of the sialic acid of the lysosomic glycoproteins facilitates the rupture of the phagosomal membrane by Tc-TOX, promoting the rapid escape of the protist from the phagosome (52).
The trypomastigotes are thus released into the cytoplasm, and in this environment, undergo a significant structural transformation (disappearance of the free flagellum and the undulating membrane, rounding of their shape, size reduction), into amastigotes. These forms will multiply by successive binary divisions (usually nine), producing from 50 to 500 protists, depending on the T. cruzi strain and the cell type involved. Approximately 12 hours prior to cell rupture, the amastigotes transform into blood trypomastigotes, which, after lysis of the infected cell, may penetrate new nucleated cells of the vertébrate host, especially smooth muscle, skeletal, cardiac, and nervous, enabling their persisting presence in the host. Due to the circulation of the blood trypomastigote forms, transfusional and congenital transmission can occur - in the latter case, related to the multiplication of the parasite in the placenta. If the vertebrate host is bitten by another triatomine, it may ingest the blood trypomastigotes (wide forms), which will reach the stomach, midgut and hindgut of the insect, transforming into epimastigotes, which multiply sequentially transforming, immediately, into metacyclic trypomastigotes (in a process called metacyclogenesis). These will adhere by the external flagellum to the surface of the hindgut wall, and part of these will be carried by the jet of excreta that drags them to the outside, allowing for their deposit on the skin surface and consequently presenting a potential infection for the source of the blood meal, and thus restarting the cycle.
Trypanosoma cruzi transcriptome analysis
Recent genetic studies have shown intraspecific differences between the genomes of the two larger strains of T. cruzi, Sylvio X10/1 and CL Brener (65). This work did not find great differences in the chief genetic content by means of analysis of presence/absence, but 6 open reading frames in CL Brener are absent in Sylvio X10/1. Many multicopy gene families are substantially reduced in Sylvio X10/1 (65). Such intraspecific difference in T. cruzi genome may generate diverging gene expression patterns, what might be used in future strategies of control of the pathogen. This difference may have epidemiologic and functional implications which must be investigated through the analysis of the genome expression of those strains. Thus the importance of integrating distinct research fields, in particular, the "omics" sciences - genomics, transcriptomics and proteomics - aiming to understand the intraspecific variability of the pathogen.
With the advances in genomic sequencing, DNA microarrays provide a powerful analytical platform for the determination of global gene expression patterns based on the quantification of mRNA levels (66). There is a variety of studies employing this methodology for the study of the abundance of transcripts in Leishmania spp. And T. brucei, but fewer studies that approach the question of the regulation of gene expression in T. cruzi. Microarray analysis of gene expression during the life cycle of these protozoa is an excellent way of identifying possible sites for drug action and the development of vaccines, taking into account the appropriate expression patterns. The initial studies on T. cruzi had a restricted global coverage - due to the absence, at the time, of a complete reference genome - using a small number of selected or random Expressed Sequence Tags (EST) or Open Reading Frame (ORF), based on genome sequencing libraries (normalized or not), to determine the relative abundance of mRNAs in the different stages of the complex life cycle of this protist (67,68). With the conclusion of the sequencing projects and in possession of complete information about the genome of these microorganisms, several research groups have invested efforts in the quantification and deduction of the transcriptome during cellular differentiation and development, using oligotide whole genome microarrays (69-73).
One of the first studies described the development of a T. cruzi DNA microarray system using a limited number and partially sequenced genomic fragments and ORFs for the identification of overexpressed genes during the transformation of trypomastigotes into amastigotes in the Brazil strain (67). This large-scale gene expression analysis identified 38 overexpressed genes in amastigotes, evidencing for the first time, genes that potentially regulate at the mRNA level the molecular events involved with the transcription stage in these microorganisms.
Overall, some previous studies of T. cruzi microarrays and those of other related protozoa of the Kinetoplastida order have suggested that for the majority of the genes of these organisms, the abundance of transcripts does not vary significantly between the life cycle stages and rarely correlates with the abundance of proteins. This last issue is very important, since it is assumed that in these organisms, transcription initiation plays a minor role in gene expression due to the generally accepted view that control is almost entirely post-transcriptions. The ORFs are organized in long arrays and are transcribed as polycistronic precursor RNAs, and are subsequently processed into individual mRNAs by trans-splicing and polyadenylation, supporting the concept that the abundance of transcripts is largely constitutive (74,75). These small variations in the abundance of mRNAs observed may result directly from the trypanosomatid biology, which have few known RNA polymerase II promoters, and may be largely incapable of controlling gene expression at the level of transcription initiation (76). However, these differences in stage-specific regulation of the relative abundance of transcripts may also likely be subject to selective mechanisms of mRNA degradation and translation via specific sequences present in the 3´- UTR regions (74,75).
An additional contribution to the debate on the usefulness of microarray studies in trypanosomatids was the overall observation in previous studies that relatively few genes exhibit significant regulation of the relative abundance of the transcriptome. For example, Koumandou, et al., using arrays targeted to the T. brucei membrane transport system (~ 10% of the genome) to compare changes in the transcriptome between the blood and procyclic promastigotes, found that only 6% of the genes were quantitatively differentially regulated between the two stages of the life cycle (71). Rochette, et al. used entire-genome oligo microarrays to compare the transcriptome of the procyclic amastigotes and promastigotes and demonstrated that only 7% of the genes in L. infantum and 9% of the genes in L. major were differentially regulated during development (72). These observations substantiate the concept that the abundance of transcripts in T. brucei and Leishmania spp. are largely constitutive throughout development. This scenario has been reinforced several times by subsequent studies, both in Leishmania spp. as well as T. brucei (69,70,77-79).
In contrast to these publications, a study of T. cruzi - using full genome microarrays - demonstrated that approximately 83% (10,256/12,288) of the oligos detected transcripts above background levels and through significance analysis of the microarrays (SAM), the relative abundance of these transcripts was significantly, differentially up or downregulated, in 50% of genes in at least one of the four stages of the T. cruzi life cycle (80). The comparison of the genes regulated in the same direction between two life stages showed that 76% (1,083/1,423) were co-induced in the developmental stages that occur within the same host (mammalian host - amastigotes and trypomastigotes; insect hosts - epimastigotes and metacyclic trypomastigotes), while roughly 20% (282/1,423) were co-induced in stages with similar biological functions. For instance, genes in amastigotes and epimastigotes (proliferative phases) were enriched in those involved in DNA repair. Likewise, the coinduced genes in tripomastigotes and metacyclic trypomastigotes (infective stages) were enriched in trans-sialidases (TS), which are involved in the invasion process (80). The differential expression for 25 of these genes was confirmed in terms of direction of expression by quantitative PCR, although there are some quantitative differences, probably as a result of differences in dynamics between the two platforms.
The reasons for the wide disparities in the degree of transcript abundance between T. cruzi and other previous studies, according to the authors, are many. First, the use of oligonucleotide probes designed with a more uniform standard for kinetic hybridization and greater genome coverage may increase the percentage of stage-regulated genes in relation to previous studies of other Kinetoplastida. According to the authors, the statistical method used in the data processing could not have contributed to the overestimation of the number of potentially regulated genes, since reanalysis of the data of Rochette, et al. (2008) (72) using their own statistical package for detection, demonstrated similar frequency percentages (7% for L. infantum) of the differentially expressed genes among amastigotes and promastigotes. Moreover, by incorporating all four phases of the T. cruzi life cycle in the analyses, contrasting with only two stages compared in the other studies, there was an increased possibility of indentifying a larger number of regulated genes in at least one of the four stages of the life cycle. Finally, because the four different stages used in the analysis were entirely differentiated, some genes excluded from the analysis may in fact be differentially expressed.
Saxena, et al. (2007) (79), in a temporal analysis L. donovani differentiation from promastigote to amastigote, demonstrated that 136 genes - about 40% of the total number of differentially regulated genes identified in the study -, are temporarily up or downregulated. Thus, it is likely that the percentage T. cruzi genes, in the present study, have been underestimated, suggesting that the temporal investigations of differentiation between the stages are important for the identification of target genes for interventions (79,80). Perhaps the most striking difference between T. cruzi and other microorganisms - T. brucei and Leishmania spp. - is that only T. cruzi has both an intracellular and an extracelular stage in vertebrate hosts, requiring a substantial genetic plasticity to quickly regulate its physiological needs in response to stress such as different environments (80). The global stageregulated gene expression data from T. cruzi are still somewhat controversial despite microarray data in accordance with the known data for protein expression for some functional groups (81-84). New studies of temporal expression during the process of transition between stages and the use of new RNA-Seq platforms in a larger scale will enable a more comprehensive understanding of the levels of regulation of gene expression and possibly the discovery of new transcripts, opening new possibilities for the development of vaccines and chemotherapy for CD (85).
Trypanosoma cruzi proteome analysis
Sequencing of the T. cruzi genome was recently completed (86); however, proteomic analysis of the different stages of T. cruzi has been performed, providing important evidence on the stage-specific expression of various proteins (87). These studies represent a useful tool for the study of the global gene expression pattern. The application of this method to trypanosomatids is particularly important, because these organisms do not use transcription initiation as a regulatory step in the control of gene expression. Several studies have demonstrated that in T. cruzi, as with other trypanosomatids, transcription occurs through polycistronics, with trans-splicing and more rarely cissplicing, and that the control of gene expression is overall performed at the post-transcriptional level, with only 10% of genes differentially expressed at the transcriptional level (88). Several lines of evidence suggest that transduction in trypanosomatids presents important differences when compared with other eukaryotes. However, little is known on the subject (88). Ayub, et al. (2009) (89) conducted a detailed search for genes of ribosomal proteins in the T. cruzi genome database together with mass spectrometry analysis of purified ribosomes from T. cruzi. The results showed that the T. cruzi ribosomal proteins share approximately 50% sequence identity with yeasts. However, some T. cruzi proteins are longer due to the presence of various N or C-terminal extensions, which are unique to trypanosomatids (89).
Proteomic analysis has proven a useful tool in the study of T. cruzi. Proteomic studies have been used to establish connections between the phylogenetic diversity of T. cruzi and quantitative patterns of protein expression (90). This way many proteins may be specifically associated to their phylogenetic subdivisions, thus being used as specific biomarkers of the pathogen subspecies (90), and helping to find the possible causes for the clinical variability of the disease. Likewise, proteomics supports the search for proteins that are expressed in only one phase of the pathogen life, therefore serving as phase markers and antigens for possible vaccines. In fact, experimental conditions for studying the proteome of trypomastigotes, amastigotes and epimastigotes by two-dimensional electrophoreses (2-DE), were optimized by Paba, et al. (2004) (91), revealing that some proteins included in the 2-DE maps exhibit significant differential expression between the three developmental forms of the parasite, resulting in 26 identifications that corresponded to expression of 19 different proteins. Among the polypeptides identified, there was expression of proteins typical of heat shock (HSP, chaperones, HSP 60, HSP 70, and HSP 90), elongation factors, glycolytic pathway enzymes (enolase, pyruvate kinase, and 2,3-biophosphoglycerate mutase) as well as structural proteins (tubulin) (91).
In this sense, it is expected at each developmental stage of T. cruzi, a specific set of proteins responsible for particular biological characteristics to be expressed. Although several articles report the quantification of T. cruzi specific mRNAs or proteins at different stages of its life cycle (92- 95), an overview of the T. cruzi proteome had not been published until 2004. The 2-D polyacramide gels for trypomastigotes, amastigotes and epimastigotes showed similar protein profiles, although differences in intensity of various spots have been observed. Each gel displayed, on average, about 500 spots in the 4-7pH range. The gels were compared in two different experiments: trypomastigotes versus amastigotes and epimastigotes versus trypomastigotes.
Comparative analysis of 2-DE gel images from the mammal forms showed that 48 spots were detected specifically in the trypomastigote, while 39 were found only in amastigotes. Furthermore, another 27 spots were significantly more intense in trypomastigotes than in amastigotes and 29 spots showed strong expression in amastigotes. When the 2-DE gels of the insect forms were compared, trypomastigotes exhibited 10 stagespecific spots and 44 more intense spots. Twenty spots were observed only in epimastigotes and 12 were more highly expressed in this form tan in other forms (91). In this study, computer analysis of the two-dimensional gel images revealed that most T. cruzi proteins present conserved expression among the three developmental forms. These results suggest that the distinct characteristics of each life cycle form of T. cruzi result from the expression of a limited number of proteins.
In accordance with what was previously mentioned, metacyclogenesis is a process that included the transformation from a non-infective epimastigote form to the infective trypomastigote form in such a way that T. cruzi acquires the capacity to penetrate into host cells. However, metacyclogenesis remains poorly understood at the molecular level. It is known that this phenomenon involves the differential expression of genes associated with the acquisition of virulence (95-97). Moreover, metacyclogenesis can be induced in vitro, so that cells in various stages of this process can be collected and their biological properties and gene expression products studied (98,99).
The metacyclic trypomastigote forms are usually involved in the establishment of infection in the vertebrate host. Studies of the molecular mechanisms of invasion led to the identification of a number of protein molecules involved in parasite-host cell interactions (100) and initiation signals that facilitate T. cruzi entry to cells (101). However, there is still a limited understanding of the infection process, and further studies are needed to characterize the complete group of proteins and molecular interactions vital to its success.
Accordingly, identification of the peptide produced by metacyclic trypomastigotes may broaden the understanding of the infection process and also allow for the description of the biochemical pathways involved in this phase, leading to the identification of target proteins for drugs and/or the design of a vaccine (102). Indeed the proteomic study of the metacyclogenesis process, performed by Parodi-Talice, et al. (2007) (102), grouped proteins into functional categories allowing for the observation that approximately 33% of the detected proteins were connected to various metabolic activities, and that the main protein classes were related to electron transport and antioxidant responses (16%), biosynthesis, catabolism, and proteolysis proteins (16%), and organization of the cytoskeleton and flagellum (12%). Other classes identified included hypothetical proteins (14%) and other peptide such as adenylate kinase, calmodulin and the surface glycoprotein GP90 (summed to 9%). The various proteins detected were attributed to different cellular compartments: the cytoplasm, nucleus, mitochondria, endoplasmic reticulum, glycossome, and cell surface (102). The expression profile of each protein identified in the 2D gel of metacyclic trypomastigotes during metacyclogenesis, was analyzed by preparing total protein extracts from five selected stages: epimastigotes, nutritionally stressed epimastigotes, differentiating (adhering) epimastigotes (3 and 24 h), and fully differentiated metacyclic trypomastigotes. The proteins were classified in three groups according to maximum expression level. Proteins such as aconitase, tiol-transferase, enolase, glutamate dehydroxygenase - which are associated with responses to different types of stress - had higher expression among the nutritionally stressed parasites; the second group consists of proteins that displayed maximum expression at differentiation after 24h; while the third group of proteins was more strongly expressed in the metacyclic form. A high number of β e α-tubulins belonging to this group are also expressed (102).
Proteomic analysis has also been proposed as an important step in the development of alternative trypanocide drugs (103,104). In the absence of effective vaccines, the control of human diseases caused by trypanosomes relies heavily on chemotherapy treatments. The current treatment for Chagas disease is based on the administration of benzonidazole and nifurtimox, which are effective against acute infections, but their use and efficacy in the chronic phase remain controversial, besides having significant side effects which make treatment more difficult (105, 106). Due primarily to the side effects and low levels of apparent cure (107), especially in the chronic phase where most cases are diagnosed (106), the need for development of new drugs for the treatment of this disease remains urgent. The most recent study seeking to identify the proteins involved in the trypanocidal activity of new compounds was performed by Menna-Barreto, et al. (2010) (108). The naphthoimidazoles tested in epimastigotes interfere with multiple mechanisms such as redox metabolism, energy production, ergosterol biosynthesis, cytoskeleton assembly, protein metabolism and biosynthesis, and chaperone modulation. These substances induce an imbalance in critical pathways of the parasite, leading to loss of metabolic homeostasis and the death of T. cruzi. Thus, proteomic approaches coupled with the development of new high-capacity technologies have been shown to be interesting methods for the discovery of good targets for chemotherapy.
Final considerations
There is still no effective treatment or vaccine against T. cruzi infection and the consequent Chagas disease. The characterization of the life cycle forms of T. cruzi, their transformations during stage changes, subproteomes and posttransductional changes generate a body of data that helps in the understanding of the biology of the parasite, identification of the proteins involved in motility, viability, and infectivity of the parasites, as well as the parasite-host interactions. It is essential that new and more modern methods of fractioning, more powerful methods of quantification be available and that in silico analysis can be improved to analyze the peculiarities of not only this but also other infectious agents. The recent discovery of T. cruzi mitocondrial DNA and its integration with the vertébrate host genome (109,110) adds new levels of complexity and challenges to understanding the host/T. cruzi interaction.
Contribution of authors
1. A. V. Martins and M. Geller proposed the structure of this article and prepared the first version. E. G. Mendonça and M. G. A. Oliveira organized the section "T. cruzi PROTEOME ANALYSIS", while J. L. R. Fietto and R F. Santos worked in the "T. cruzi TRANSCRIPTOME ANALYSIS" and in the figure 1 and figure 2. A. P. Gomes, L. A. Santana, R. R. Vitorino, R. Siqueira-Batista worked in the process of literatura review and critical revision of the text.
Correspondencia: Prof. Dr. Rodrigo Siqueira Batista, Departamento de Medicina e Enfermagem, Universidade Federal de Viçosa. Avenida P. H. Rolfs s/n, Campus Universitário, Viçosa, MG, Brasil, CEP 36571-000 Dirección electrónica: rsbatista@ufv.br
References
1. Amato Neto V, Pasternak J. Centenário da doença de Chagas. Ver Saúde Pública. 2009;43:381-2. [ Links ]
2. Castro I, Andrade JP, Paola AAV, Vilas-Boas F, Oliveira GMM, Marin Neto JA. I Diretriz Latino-Americana para o Diagnóstico e Tratamento da Cardiopatia Chagásica. Arq Bras Cardiol. 2011;97(Supl.3):1-48. [ Links ]
3. Pittella JEH. O processo de avaliação em ciência e a indicação de Carlos Chagas ao prêmio Nobel de Fisiologia ou Medicina. Rev Soc Bras Med Trop. 2009;42:67-72. [ Links ]
4. Cabrera R, Veja S, Cáceres AG, Ramal AC, Álvarez C, Ladeira P, et al. Epidemiological investigation of na acute case of Chagas disease in an area of active tramsission in Peruvian amazon region. Rev Inst Med Trop. 2010;52:269-72. [ Links ]
5. Dias JCP. Doença de Chagas, ambiente, participação e Estado. Cad Saúde Pública. 2001;17(Supl.1):165-169. [ Links ]
6. Maguire JH. Chagas´ disease - can we stop the deaths? N Engl J Med. 2006;355:760-1. [ Links ]
7. Tanowitz HB, Machado FS, Jelicks LA, Shirani J, de Carvalho AC, Spray DC, et al. Perspectives on Trypanosoma cruzi-induced heart disease (Chagas disease). Prog Cardiovasc Dis. 2009;51:524-39. [ Links ]
8. Bonney KM, Engman DM. Chagas heart disease pathogenesis: one mechanism or many? Curr Mol Med. 2008;8:510-8. [ Links ]
9. Rochitte CE, Nacif MS, Júnior Oliveira AC, Siqueira-Batista R, Marchiori E, Uellendahl M, et al. Cardiac magnetic resonance in Chagas´ disease. Artif Organs. 2007;31:259-67. [ Links ]
10. Dhiman M, Nakayasu ES, Madaiah YH, Reynolds BK, Wen JJ, Almeida IC, et al. Enhanced nitrosative stress during Trypanosoma cruzi infection causes nitrotyrosine modification of host proteins. AJP. 2008;173:728-40. [ Links ]
11. Capriles PV, Guimarães AC, Otto TD, Miranda AB, Dardenne LE, Degrave WM. Structural modelling and comparative analysis of homologous, analogous and specific proteins from Trypanosoma cruzi versus Homo sapiens: putative drug targets for Chagas´ disease treatment. BMC Genomics. 2010;11:610. [ Links ]
12. Villalta F, Scharfstein J, Ashton AW, Tyler KM, Guan F, Mukherjee S, et al. Perspectives on the Trypanosoma cruzi-host cell receptor interactions. Parasitol Res. 2009;104:1251-60. [ Links ]
13. Schijman AG, Bisio M, Orellana L, Sued M, Duffy T, Mejia Jaramillo AM, et al. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis. 2011;5:e931. [ Links ]
14. Sposito AC, Ramires JAF. Doença de Chagas: avanços recentes no diagnóstico e tratamento. In: Fauci AS, Braunwald E, Kasper DL, Hauser SL, Longo J, Jameson L, et al (editores). Harrison Medicina Interna. 18a edição. Rio de Janeiro: McGraw-Hill; 2008. [ Links ]
15. Roberts SB, Robichaux JL, Chavali AK, Manque PA, Lee V, Lara AM, et al. Proteomic and network analysis characterize stage-specific metabolism in Trypanosoma cruzi. BMC Systems Biology. 2009;3:52. [ Links ]
16. Hoare CA. Rationalization of the terminology for the developmental stages of trypanosomatid flagellates. Med Parazitol (Mosk). 1971;40:307-9. [ Links ]
17. Mattei DM, Goldenberg S, Morel C. Biochemical strain caracterization of Trypanosoma cruzi by restriction endonuclease cleavage of kinetoplast-DNA. FEBS Letters. 1977;74:264-8. [ Links ]
18. Nogueira N, Coura JR. American Trypanosomisasis (Chagas´ Disease). In: Warren KS, Mahmoud ADF (editors). Tropical and Geographical Medicine. 2nd edition. New York: Mc-Gran Hill; 1989. [ Links ]
19. Rey L. Parasitologia. 4a ed. Rio de Janeiro: Guanabara Koogan; 2008. [ Links ]
20. Brener Z. Intraspecific variations in Trypanosoma cruzi: two types of parasite populations presenting distinct characteristics. PAHO. 1997;347:11-21. [ Links ]
21. Levine ND, Corliss JO, Cox FEG. A newly revised classification of the protozoa. J Protozool. 1980;27:37-58. [ Links ]
22. Mccabe RE, Meagher S, Mullins B. Trypanosoma cruzi: Explant organ cultures from mice with Chagas´ disease. Exp Parasitol. 1989; 68:462-9. [ Links ]
23. Tibayrenc M. Population genetics of parasitic protozoa and other microorganisms. Adv Parasitol. 1995;36:48-115. [ Links ]
24. Andrade SG, Magalhães JB. Biodemes and zymodemes of Trypanosoma cruzi strains: correlations with clinical data and experimental pathology. Rev Soc Bras Med Trop. 1997;30:27-35. [ Links ]
25. Zingales B, Stolf BS, Souto RP, Fernandes O, Briones MR. Epidemiology, biochemistry and evolution of Trypanosoma cruzi lineages based on ribosomal RNA sequences. Mem Inst Oswaldo Cruz. 1999;94:159-64. [ Links ]
26. Martinez-Diaz RA, Escario JA, Nogal-Ruiz JJ, Gomez-Barrio A. Biological characterization of Trypanosoma cruzi strains. Mem Inst Oswaldo Cruz. 2001;96:53-9. [ Links ]
27. Andrade SG. Biodemas, zimodemas e esquizodemas: sua relação com a Patologia da doença de Chagas. In: Coura JR (editor). Dinâmica das Doenças Infecciosas e Parasitárias. Rio de Janeiro: Guanabara Koogan; 2005. [ Links ]
28. Siqueira-Batista R, Geller M, Martins AV, Bastos OMP. Trypanosoma cruzi. In: Siqueira-Batista R, Gomes AP, Corrêa AD, Geller M (editores.). Moléstia de Chagas. 2ª edição. Rio de Janeiro: Rúbio; 2007. [ Links ]
29. Momem H. Taxonomy of Trypanosoma cruzi: a commentary on characterization and nomenclature. Mem Inst Oswaldo Cruz. 1999;94(Suppl.1):181-4. [ Links ]
30. Chiari E, Galvão LMC, Lages-Silva E. Diagnóstico parasitológico e caracterização biológica, bioquímica e genética de tripanosomas parasitos do homemm. In: Coura JR (editor). Dinâmica das Doenças Infecciosas e Parasitárias. Rio de Janeiro: Guanabara Koogan; 2005. [ Links ]
31. Villela MM, Souza JMB, Melo VP, Dias JCP. Vigilância epidemiológica da doença de Chagas em programa descentralizado: avaliação de conhecimentos e práticas de agentes municipais em região endémica de Minas Gerais, Brasil. Cad. Saúde Pública. 2007;23:2428-38. [ Links ]
32. Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, et al. Second Satellite Meeting. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009;104:1051-4. [ Links ]
33. Andrade SG, Figueira RM, Carvalho ML, Gorini DF. Influência da cepa do Trypanosoma cruzi na resposta à terapêutica experimental pelo Bay 2502. Rev Inst Med Trop São Paulo. 1975;18:140-1. [ Links ]
34. Brener Z. Present status of chemotherapy and chemoprophylaxis of human trypanosomes in the western hemisphere. Pharmacol Therap. 1979;7:71-90. [ Links ]
35. Dantas AP, Barbosa HS, de Castro SL. Biological and ultrastructural effects of the anti-microtubule agent taxol against Trypanosoma cruzi. J Submicrosc Cytol Pathol. 2003;35:287-94. [ Links ]
36. Brener Z. Trypanosoma cruzi: morfologia e ciclo evolutivo. Dias JCP, Coura JR (editores). In: Clínica e Terapêutica da doença de Chagas: uma abordagem prática para o clínico geral. Rio de Janeiro: FIOCRUZ; 1997. [ Links ]
37. Adade CM, de Castro SL, Soares MJ. Ultrastructural localization of Trypanosoma cruzi lysosomes by aryl sulphatase cytochemistry. Micron. 2006;38:252-6. [ Links ]
38. De Souza W. Special organelles of some pathogenic protozoa. Pa rasitol Res. 2002;88:1013-25. [ Links ]
39. Dias JCP, Macedo VO. Doença de Chagas. In: Coura JR (editor). Dinâmica das Doenças Infecciosas e Parasitárias. Rio de Janeiro: Guanabara Koogan; 2005. [ Links ]
40. Borst P, Hoeijmakers JHJ. Structure and function of kinetoplast DNA of African trypanosomes. In: Cummings DP. Borst I, David S, Weissman, Fox CF (editors). Extrachromosomal DNA ICN-UCLA Symposia on Molecular and Cellular Biology. New York: Academic Press; 1979. [ Links ]
41. Fairlarm AH, Weislogel PO, Hoeijmakers JHJ, Borst P. Isolation and characterization of kinetoplast DNA from bloodstream from of Trypanosoma brucei. J Cell Biol. 1978;76:293-309. [ Links ]
42. Kleisen CM, Borst P. Sequence heterogeneity of the minicircles of kinetoplast DNA of Crithidia luciliae and evidence for the presence of a component more complex than mini-circle DNA in the kinetoplast network. Biochim Biophys Acta. 1975;407:473-8. [ Links ]
43. Kleisen CM, Weislogel PO, Fonk K, Borst P. The structure of kinetoplast DNA. I- The mini-circles of Crithidia luciliae are heterogeneous in base sequence. Eur J Biochem. 1976;64:144-51. [ Links ]
44. Degrave W, Fragoso SP, Britto C, Van Heuverswyn H, Kidane GZ, Cardoso MAB, et al. Peculiar sequence organization of kinetoplast DNA minicircles from Trypanosoma cruzi. Mol Biochem Parasitol. 1988;27:63-70. [ Links ]
45. Morel CM. Kinetoplast DNA minicircles as multipurpose molecular markers for Trypanosoma cruzi characterization and detection. In: Schwartz M (editor). Molecular biology and infectious diseases (Colloque du Centenaire de L´Institut Pasteur), Paris: Elsevier; 1988. [ Links ]
46. Macina RA, Sanchez D, Gluschankof D, Burrone O, Frascha ACC. Sequence diversity in the kinetoplast DNA minicircles of Trypanosoma cruzi. Mol Biochem Parasitol. 1986;21:25-32. [ Links ]
47. Weslei RD, Simpson L. Studies in kinetoplast DNA. III - Kinetic complexing of kinetoplast and nuclear DNA from Leishmania tarentolae. Biochim Biophys Acta. 1973;319:267-80. [ Links ]
48. Pollard VW, Rohrer SP, Michlotti EF, Hancock K, Hajduk SL. Organization of minicircle genes for guide RNAs in Trypanosoma brucei. Cell. 1990;63:783-90. [ Links ]
49. Sturm NR, Simpson L. Leishmania tarentolae minicircles of different sequence classes encode single guide RNAs located in the variable region approximately 150 bp from the conserved region. Nucl Acids Res. 1991;19:6277-81. [ Links ]
50. Souza W. Electron microscopy of trypanosomes - a historical view. Mem Inst Oswaldo Cruz. 2008;103:313-25. [ Links ]
51. Landfear SM, Ignatushchenko M. The flagellum and flagellar pocket of trypanosomatids. Mol Biochem Parasitol. 2001;115:1-17. [ Links ]
52. Field MC, Carrington M. The trypanosome flagellar pocket. Nat Rev Microbiol. 2009;7:775-86. [ Links ]
53. Docampo R, de Souza W, Miranda K, Rohloff P, Moreno SN. Acidocalcisomes - conserved from bacteria to man. Nat Rev Microbiol. 2005;3:251-61. [ Links ]
54. Lopez-Velazquez G, Hernandez R, Lopez-Villasenor I, Reyes-Vivas H, Segura-Valdez MDEL, Jimenez-Garcia LF. Electron microscopy analysis of the nucleolus of Trypanosoma cruzi. Microsc Microanal. 2005;11:293-9. [ Links ]
55. Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br Med Bull. 1985;41:105-14. [ Links ]
56. Corrêa AD, Miranda Filho N, Siqueira-Batista R, Huggins DW. Papel dos reservatórios na epidemiologia da moléstia de Chagas. Rev Bras Med. 1998;55:414-20. [ Links ]
57. Hoare CA, Wallace FG. Developmental stages of trypanosomatid flagellates: A new terminology. Nature. 1966;212:1385-6. [ Links ]
58. Brener Z. Biology of Trypanosoma cruzi. Ann Rev Microbiol. 1973;27:347-82. [ Links ]
59. Bourguignon SC, Mello CB, Gonzalez MS, Souto-Padron T. Biological aspects of the Trypanosoma cruzi (Dm28c clone) intermediate form, between epimastigote and trypomastigote, obtained in modified liver infusion tryptose (LIT) medium. Acta Trop. 2006;98:103- 92. [ Links ]
60. Hoare CA. The Trypanosomes of Mammals: A Zoological Monograph. Oxford: Blackwell; 1972. [ Links ]
61. Ley V, Andrews NW, Robbins ES, Nussenzweig V. Amastigotes of Trypanosoma cruzi sustain an infective cycle in mammalian cells. J Exp Med. 1988;168:649-9. [ Links ]
62. Florencio-Martínez L, Márquez-Dueñas C, Ballesteros-Rodea G, Martínez-Calvillo S, Manning-Cela R. Cellular analysis of host cell infection by different developmental stages of Trypanosoma cruzi. Exp Parasitol. 2010;126:332-6. [ Links ]
63. Alvarenga NJ, Bronfen E. Metaciclogênege do Trypanosoma cruzi como parâmetro de interação do parasita com o triatomíneo vetor. Rev Bras Med Trop. 1997;30:247-50. [ Links ]
64. World Health Organization. Control of Chagas Disease. Second Report of the WHO Expert Committee: Technical Report Series. Geneva: World Health Organization; 2002. [ Links ]
65. Franzén O, Ochaya S, Sherwood E, Lewis MD, Llewellyn MS, Miles MA, et al. Shotgun Sequencing Analysis of Trypanosoma cruzi I Sylvio X10/1 and Comparison with T. cruzi VI CL Brener. PLoS Negl Trop Dis. 2011;5: e984. [ Links ]
66. Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nature Genet. 1999;21:33-7. [ Links ]
67. Minning TA, Bua J, Garcia GA, McGraw RA, Tarleton RL. Microarray profiling of gene expression during trypomastigote to amastigote transition in Trypanosoma cruzi. Mol Biochem Parasitol. 2003;131:55-64. [ Links ]
68. Baptista CS, Vêncio RZN, Abdala S, Valadares MP, Martins C, de Bragança Pereira CA, et al. DNA microarrays for comparative genomics and analysis of gene expression in Trypanosoma cruzi. Mol Biochem Parasitol. 2004:138:183-94. [ Links ]
69. Holzer TR, McMaster WR, Forney JD. Expression profiling by wholegenome interspecies microarray hybridization reveals differential gene expression in procyclic promastigotes, lesion-derived amastigotes, and axenic amastigotes in Leishmania mexicana. Mol Biochem Parasitol. 2006;146:198-218. [ Links ]
70. Cohen-Freue G, Holzer TR, Forney JD, McMaster WR. Global gene expression in Leishmania. Int J Parasitol. 2007;37:1077-86. [ Links ]
71. Koumandou VL, Natesan SKA, Sergeenko T, Field MC. The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics. 2008;9:298. [ Links ]
72. Rochette A, Raymond F, Jean-Michel U, Smith M, Messier N, Boisvert S, et al. Genome-wide gene expression profiling analysis of Leishmania major and Leishmania infantum developmental stages reveals substantial differences between the two species. BMC Genomics. 2008;9:255. [ Links ]
73. Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nature Genetics. 1999;21(Suppl.1):33-7. [ Links ]
74. Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156:93-101. [ Links ]
75. Haile S, Papadopoulou B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr Opin Microbiol. 2007;10:569-77. [ Links ]
76. Campbell DA, Thomas S, Sturm NR. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect. 2003;5:1231-40. [ Links ]
77. Diehl, S., F. Diehl, El-Sayed NM, Clayton C, Hoheisel JD. Analysis of stage-specific gene expression in the bloodstream and the procyclic form of Trypanosoma brucei using a genomic DNA-microarray. Mol Biochem Parasitol. 2002;123:115-23. [ Links ]
78. Luu V D, Brems S, Hoheisel JD, Burchmore R, Guilbride DL, Clayton C. Functional analysis of Trypanosoma brucei PUF1. Mol Biochem Parasitol. 2006;150:340-9. [ Links ]
79. Saxena A, Lahav T, Holland N, Aggarwal G, Anupama A, Huang Y, et al. Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Mol Biochem Parasitol. 2007;152:53-65. [ Links ]
80. Minning TA, Weatherly DB, Atwood J 3rd, Orlando R, Tarleton RL. The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genomics. 2009;10:370. [ Links ]
81. Paba J, Santana JM, Teixeira AR, Fontes W, Sousa MV, Ricart CA. Proteomic analysis of the human pathogen Trypanosoma cruzi. Proteomics. 2004;4:1052-9. [ Links ]
82. Parodi-Talice A, Durán R, Arrambide N, Prieto V, Piñeyro MD, Pritsch O, et al. Proteome analysis of the causative agent of Chagas disease: Trypanosoma cruzi. Int J Parasitol. 2004;34:881-6. [ Links ]
83. Atwood 3rd JA, Weatherly DB, Minning TA, Bundy B, Cavola C,Opperdoes FR, et al. The Trypanosoma cruzi proteome. Science. 2005;309:473-6. [ Links ]
84. Atwood JA 3rd, Minning T, Ludolf F, Nuccio A, Weatherly DB, Alvarez-Manilla G, et al. Glycoproteomics of Trypanosoma cruzi trypomastigotes using subcellular fractionation, lectin affinity, and stable isotope labeling. J Proteome Res. 2006; 59:3376-84. [ Links ]
85. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57-63. [ Links ]
86. El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409-15. [ Links ]
87. Leifso K, Cohen-Freue G, Dogra N, Murray A, McMaster WR. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: the Leishmania genome is constitutively expressed. Mol Biochem Parasitol. 2007;152:35-46. [ Links ]
88. Martínez-Calvillo S, Vizuet-de-Rueda JC, Florencio-Martínez LE, Manning-Cela RG, Figueroa-Angulo EE. Gene expression in trypanosomatid parasites. J Biomed Biotechnol. 2010; Epub 2010 Feb 11. [ Links ]
89. Ayub MJ, Atwood J, Nuccio A, Tarleton R, Levin MJ. Proteomic analysis of the Trypanosoma cruzi ribosomal proteins. Biochem Biophys Res Commun. 2009; 382:30-4. [ Links ]
90. Telleria J, Biron DG, Brizard JP, Demettre E, Seveno M, Barnabé C, et al. Phylogenetic character mapping of proteomic diversity shows high correlation with subspecific phylogenetic diversity in Trypanosoma cruzi. Proc Natl Acad Sci USA. 2010;107:20411-6. [ Links ]
91. Cuervo P, Domont GB, Jesus JB. Proteomics of trypanosomatids of human medical importance. J Proteomics. 2010;73:845-67. [ Links ]
92. Teixeira MGM, Borges-Pereira J, Netizert E, Souza MLNX, Peralta JM. Development and evaluation of an enzyme-linked immunotransfer blot technique for serodiagnosis of Chagas disease. Trop Med Parasitol. 1994;45:308-12. [ Links ]
93. Freitas-Júnior LHG, Briones MRS, Schenkman S. Two distincts groups of mucin-like genes are differentialy expressed in the developmental stages of Trypanosoma cruzi. Mol Biochem Parasitol. 1998;93:101-14. [ Links ]
94. Grandgenett PM, Coughlin BC, Kirchhoff LV, Donelson JE. Differential expression of GP63 genes in Trypanosoma cruzi. Mol Biochem Parasitol. 2000;110:409-15. [ Links ]
95. Ávila AR, Yamada-Ogatta SF, Silva-Monteiro V, Krieger MA, Nakamura CV, De Souza W, et al. Cloning and characterization of the metacyclogenin gene, which is specifically expressed during Trypanosoma cruzi metacyclogenesis. Mol Biochem Parasitol. 2001;117:169-77. [ Links ]
96. Contreras VT, Morel CM, Goldenberg S. Stage specific gene expression precedes morphological changes during Trypanosoma cruzi metacyclogenesis. Mol Biochem Parasitol. 1985;14:83-96. [ Links ]
97. Goldenberg S, Salles JM, Contreras VT, Lima-Franco MP, Katzin AM, Colli W, et al. Characterization of messenger RNA from epimastigotes and metacyclic trypomastigotes of Trypanosoma cruzi. FEBS Lett. 1985;180:265-70. [ Links ]
98. Krieger MA, Avila AR, Ogatta SF, Plazanet-Menut C, Goldenberg S. Differential gene expression during Trypanosoma cruzi metacyclogenesis. Mem Inst Oswaldo Cruz. 1999;94:165-8. [ Links ]
99. Ávila AR, Dallagiovanna B, Yamada-Ogatta SF, Monteiro-Goes V, Fragoso SP, Krieger MA, et al. Stage-specific gene expression during Trypanosoma cruzi metacyclogenesis. Genet Mol Res. 2003;2:159-68. [ Links ]
100. Neira I, Silva FA, Cortez M, Yoshida N. Involvement of Trypanosoma cruzi metacyclic trypomastigote surface molecule gp82 in adhesión to gastric mucin and invasion of epithelial cells Infect Immun. 2003;71:557-61. [ Links ]
101. Andrews NW. Lysosomes and the plasma membrane: trypanosomes reveal a secret relationship. J Biol Chem. 2002;158:389-94. [ Links ]
102. Parodi-Talice A, Monteiro-Goes V, Arrambide N, Avila AR, Duran R, Correa A, et al. Proteomic analysis of metacyclic trypomastigotes undergoing Trypanosoma cruzi metacyclogenesis. J Mass Spectrom. 2007;42:1422-32. [ Links ]
103. d e Souza, W. From the cell biology to the development of new chemotherapeutic approaches against trypanosomatids: dreams and reality. Kinetoplastid Biol Dis. 2002;1:3. [ Links ]
104. Soeiro MN, De Castro SL. Trypanosoma cruzi targets for new chemotherapeutic approaches Expert Opin Ther Targets. 2009;13:105- 21. [ Links ]
105. Coura JR, De Castro SL. A critical review on Chagas´ disease chemotherapy. Mem Inst Oswaldo Cruz. 2002;97:3-24. [ Links ]
106. Guedes PMM, Fietto JLR, Lana M, Bahia, M T. Advances in Chagas Disease Chemotherapy. Anti-Infective Agents. In: Medicinal Chemistry (Formerly Current Medicinal Chemistry - Anti-Infective Agents); 2006. [ Links ]
107. Jannin J, Villa L. An overview of Chagas´ disease treatment. Mem Inst Oswaldo Cruz. 2007;102(Suppl.1):95-7. [ Links ]
108. Menna-Barreto RFS, Beghini DG, Ferreira ATS, Pinto AV, De Castro SL, Perales J. A proteomic analysis of the mechanism of action of naphthoimidazoles in Trypanosoma cruzi epimastigotes in vitro. J Proteomics. 2010;73:2306-15. [ Links ]
109. Nitz N, Gomes C, De Cassia RA, D´Souza-Ault MR, Moreno F, Lauria-Pires L, et al. Heritable integration of kDNA minicircle sequences from Trypanosoma cruzi into the avian genome: insights into human Chagas´ disease. Cell. 2004;118:175-86. [ Links ]
110. Hecht MM, Nitz N, Araujo PF, Sousa AO, Rosa AC, Gomes DA, et al. Inheritance of DNA transferred from American trypanosomes to human hosts. PLoS One. 2010;5:e9181. [ Links ]














