Introduction
Patients receiving hemodialysis are highly susceptible to healthcare-associated infections, not only because this procedure is invasive, but also because of the immunocompromise caused by the uremia and the inflammation, which can cause oxidative stress and trigger apoptosis, resulting in a decrease in the number of T lymphocytes1,2. The most frequent infections are bacteremia and local infection at the place of vascular access, however other types of infections can present, such as pneumonia, endocarditis, skin and soft tissues infections and urinary tract infections3.
Bacteremia is the most frequent cause of morbidity and, after cardiovascular disease, the second most frequent cause of mortality, mainly due to catheter contamination, either in the endoluminal region as a result of manipulation or extraluminal contamination ocassioned by the migration of the body’s microbiota through the fibrin skin path around the device3. Infections also occur in about 3-10% of cases by hematogenous route from another point of infection or by the contamination of infusion fluids2. The risk of bacteremia is up to 10 times higher with catheter use compared to that of arteriovenous fistula use, occurring in up to 32% of patients treated using this device4. Likewise, patients on hemodialysis are a group at high risk of infection by resistant bacteria due to frequent hospitalizations and the history of antibiotic use, which considerably complicates the prognosis compared to infections caused by susceptible microorganisms5. The emergence of resistance to beta-lactams antibiotics has been particularly notable in Gram-positive bacteria such as Staphylococcus aureus and in Gram-negative bacilli such as Escherichia coli and Klebsiella pneumoniae; microorganisms that frequently affect the hemodialysis population5.
The objective of this review is to describe the importance of infections caused by multidrug-resistant bacteria in hemodialysis patients and the colonization as a risk factor associated with these outcomes.
Collection of information
The information used in this review was collected from three bibliographic databases: Medline, Embase and Scielo, and articles in Spanish or English that were preferably published between the years 2005-2015 were selected. The descriptors used in the search combined terms related to (i) exposures such as colonization, multidrug-resistant bacteria, Staphylococcus aureus, Gram-negative bacteria and Klebsiella pneumoniae; (ii) outcomes such as infections, bacteremia and bloodstream infections and (iii) relevant population (hemodialysis patients).
The selected studies included cross-sectional, cohort, casecontrol systematic reviews and meta-analyses. The selection of articles was focused on those whose study population consisted of hemodialysis patients. However, other studies that provide a context for the behavior of the resistance at the national level were included.
Multidrug-resistant bacteria in hemodialysis patients
Staphylococcus aureus is the most frequently found bacteria in hemodialysis patients, so the emergence of methicillin resistance has had a significant impact on this population6,7. Methicillin-resistant S. aureus isolates (MRSA) cause more than 51% of blood infections, which are associated with a significant increase in costs and mortality5. About 90% of patients with invasive MRSA infections require hospitalization and about 17% of these die during hospital stay8. When healthcareassociated infections caused by MRSA are taken into account, the incidence is significantly greater in dialysis patients compared with other groups of patients exposed to the hospital environment5. Community-associated MRSA isolates (CAMRSA) cause about 25% of infections and are the main cause of bacteremia and skin and soft tissue infections6,7.
On the other hand, Gram-negative bacilli cause around 25% of bacteremia, being approximately 9.7% resistant to thirdgeneration cephalosporins and 3.4% resistant to carbapenems9. Mortality due to these microorganisms is around 25.3%, being higher in infections caused by resistant isolates and in polymicrobial infections, which can occur in 11.9% of cases10. The common sources of bacteremia are: central venous catheters (16.8%), infected ulcers (14.1%), urinary tract (10.5%), biliary tract (9.5%) and intra-abdominal infections (9.5%), with bacteria such as Escherichia coli (49.5%), Enterobacter spp (13.1%), Klebsiella spp (11.1%), Proteus mirabilis (6.1%) and Pseudomonas aeruginosa (5.1%)10.
There are few studies that have evaluated infections caused by resistant bacteria in hemodialysis patients5; however, hemodialysis has been found to be an independent risk factor for extended-spectrum beta-lactamase-(ESBL) producing Gram negative bacilli infections, so these patients are at higher risk of infection by these bacteria compared to susceptible isolates11. It has also been reported that isolates of K. pneumoniae which produce these beta-lactamases can cause up to 23.1% of the infections that occur in hemodialysis patients12.
Factors associated with infections by multidrugresistant bacteria
The risk factors for infection by resistant bacteria in hemodialysis patients include factors specific of the individual and factors associated with healthcare, for example contact with the hospital environment and with the renal unit. Thus, a greater risk of infection has been detected in patients over 50 or 60 years old (HR: 1.03, 95% CI 1.02-1.05, p <0.001), with previous use of antibiotics (OR: 2.53, 95% CI 1.21-5.28, p = 0.013), previous bacterial infections (OR: 2.00, 95% CI 1.47- 2.72), history of hospitalization (OR: 5.90, 95% CI 1.91-18.24, p=0.002) or immunosuppressive drugs (OR: 3.0 95% CI 1.0- 6.1)13-15. As well as the presence of diseases such as diabetes (OR: 2.37, 95% CI 1.65-3.39), a low level of albumin (HR: 0.32, 95% CI 0.25-0.41, p <0.0001), and the Charlson index ( OR: 3.6, 95% CI: 1.2-10.7, p = 0.02) have been reported as predictors of these infections16,17. Some characteristics related to the dialysis process, such as the presence of a central venous catheter as opposed to a fistula (RR = 3.38, 95% CI 1.89-6.30) and a short time on dialysis (HR: 0.70, 95% CI 0.56-0.89; p = 0.003) have also been reported as risk factors13,18.
Colonization by multidrug-resistant bacteria as a risk factor for infection
Previous studies have indicated that bacterial colonization is one of the main risk factors for infection in the dialysis population19-22(Table 1). It has even been reported that the percentage of colonization by at least one multidrug-resistant bacteria in patients in the hemodialysis modality can reach 28%23. This has several implications: first because in order to prevent infections by these bacteria, strategies should be focused on avoiding transmission; second because colonization is more frequent than infection and can persist for long periods of time and third because asymptomatic carriers of multidrug-resistant bacteria are reservoirs of these microorganisms and can transmit them without being detected5.
Table 1 Studies that have evaluated colonization as a risk factor for infection by Staphylococcus aureus in patients on hemodialysis
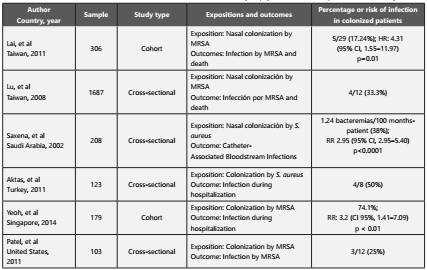
Staphylococcus aureus (particularly MRSA) is the bacteria that most frequently colonizes patients on hemodialysis, and has been reported with a percentage ranging from 1.4% to 27% of patients, of which around 17% to 35% can develop infections like bacteremia5. Colonization by this bacteria can be persistent (over long periods of time) or intermittent (with changing patterns over time), the former being associated with an increased risk of infection and increased mortality15. Transmission of MRSA can be patient to patient, through the contaminated hands or gloves of health care workers, or from contaminated surfaces, suggesting an exogenous source of transmission, with an incidence of 1.2 per 100 patient-months of risk7,23,24. For the assessment of MRSA colonization, it has been reported that screening in nostrils has a sensitivity of only 84%, which makes it necessary to evaluate other sites such as the skin and the pharynx in order to increase the sensitivity of detection to above 90%25. Previous studies suggest that skin colonization by bacteria such as MRSA may encourage the presence in skin, vascular access and blood infections21,22. Unlike MRSA, few studies have evaluated colonization by multidrug-resistant Gram-negative bacilli in dialysis patients, so their role in the development of infection has not yet been established. Previous studies have reported an intestinal colonization of 16% by these bacteria, and some authors have even suggested that this percentage may be higher compared to colonization by MRSA5,7,23. In uncolonized patients, the probability of colonization during a 6-month follow-up period can be up to 20%, either by exogenous transmission (through patient to patient) or endogenously (through the acquisition of mechanisms of resistance in bacteria that are part of the microbiota, horizontally by conjugation or vertically by clonal dissemination)7,23. Among the betalactamases that have been described are those of the ESBL type in bacteria such as Escherichia coli, at a percentage of around 8.3%26.
The gastrointestinal tract is one of the most likely sites to be colonized by multidrug-resistant bacilli (MDR-BGN), not only because of the ease in oral-fecal transmission but also because many antibiotics are eliminated by the intestinal tract21,22,27. The intestine is a center of horizontal transfer of resistance mechanisms through genetic elements from one bacteria to another and from there the translocation of these microorganisms can be presented through the bloodstream or fecal contamination of the vascular access can also occur27,28. Although the intestinal microbiota is stable and similar in members of the same family, it can be altered the use of antibiotics which can cause changes in microbial densities that can last up to four years29.
Different studies indicate that the community could be an important source of transmission of resistant bacteria, leading to colonization and subsequent infections in different types of patients, including patients on hemodialysis. The frequent use of non-prescribed antibiotics in daily life and in environments other than human health, such as veterinary medicine, agriculture and the food industry (where they are used up to four times as much), has contributed to the colonization and dissemination of resistant bacteria in the community30-33. Antibiotics are some of the most frequently sold drugs in pharmacies, often without medical formulas and for infections of viral origin, on which they have no effect34,35.
Additional factors to the consumption of antibiotics, such as stay in places of prolonged care like nursing homes, previous contact with health care services, travels to endemic areas and certain eating habits such as the consumption of pigs and poultry, have been nassociated with the acquisition of betalactmases of the ESBL type in the community; suggesting transmission through the food chain36-38.
Likewise, ESBL-producing strains of E. coli with the same genetic profile have been found on household surfaces, in pets and in humans, suggesting cross-transmission of this bacteria39. Person-to-person transmission, even from mother to child during of delivery, has been documented by different authors40,39,41, in addition to other sources of transmission such as money, water and domestic animals42-45. Colonization by bacteria such as ESBL-producing E. coli has been associated with community-acquired infections, mainly by CTX-M-1 carrier isolates, unlike hospital isolates commonly producing CTX-M-14 or CTX-1537,38,46.
In relation to contact during health care, recent hospitalization has been described by several authors as a risk factor for colonization by resistant bacteria (OR = 1.93, 95% CI, 1.04-3.58), which could be related to the acquisition of these microorganisms in the hospital environment47,48,49.
Studies that have evaluated the association between colonization and the development of infections by resistant bacteria in hemodialysis patients have limitations, such as the lack of use of molecular methods to confirm that the colonizing species is the same as that which causes infection, the measurement of exposure only at the beginning of the study, the absence of follow-up, the focus only on MRSA without including other resistant bacteria of clinical importance such as MDR-BGN, the evaluation of bacteremia as the only important outcome and the lack of knowledge about the prognosis of infections in previously colonized patients19,24,50-52.
Similarly, other authors have questioned the existence of a causal association between colonization and infection in hemodialysis patients, given that colonization may only be a marker of other patient health conditions that may make them more susceptible to infection, for example the presence of different comorbidities, the consumption of antibiotics or immunosuppressive drugs, and frequent contact with health care53. This situation has led to the fact that prophylactic decolonization is a controversial issue in terms of its effectiveness in reducing infections, despite its recommendation in national guidelines, and it is not used by healthcare personnel due to the high percentages of re-infection, the development of resistance to the antibiotics used, the selection of appropriate site for application and lack of consensus as to the dose and time of therapy53,54.
Prevention of infections in hemodialysis patients
The recommendations given by the CDC (Centers for Disease Control and Prevention) to prevent the transmission of resistant bacteria are general and not specific to multidrug-resistant bacteria in hemodialysis patients55. These recommendations are mainly based on avoiding transmission mainly by the use of gloves and gowns by care personnel, the cleaning of equipment and surfaces, the preparation of medicines in clean areas, the optimization of antibiotics and the education of patients and personnel55. However, given the complexity of the problem of antimicrobial resistance, it is necessary to implement other strategies that achieve a more effective control of the problem5.
Some of these strategies consist of the de-escalation of antibiotics such as vancomycin (used inappropriately as a targeted treatment in more than 10% of cases), the use of fistulas instead of catheters where possible, contact precautions in colonized or infected patients and decolonization5,56. The use of this last strategy is controversial because, despite evidence indicating the reduction of invasive infections by S. aureus, it may increase the risk of developing resistance57. Additionally, knowledge about antibiotics and the duration and frequency of treatment is limited5,56. Despite these strategies of prevention, the percentages of infection by resistant bacteria in hemodialysis patients continues in increasing, which leads to the need to identify other risk factors that can be intervened to prevent the transmission of these microorganisms57.
Molecular biology offers a set of typing techniques that allow more precise knowledge of the transmission patterns of resistant bacteria in a specific place, such as pulsed-field gel electrophoresis, multi-locus sequence typing and whole genome sequencing57. These offe advantages over other epidemiological tools in that they can shed light on how these microorganisms are transmitted57.
The integration of molecular biology techniques with traditional epidemiology has given rise to a branch of epidemiology called “molecular epidemiology”, which aims to define the distribution of diseases and identify risk factors in populations58,59. Molecular epidemiology has emerged as a useful tool for the evaluation of the transmission of resistant bacteria in healthcare institutions because it allows a more accurate evaluation of the behavior of infectious diseases and refines the analysis of the associations with a greater causal inference57,58. This is done through the identification of genotypes of clinical importance, the knowledge of their distribution in time, person and place and the comparison of the genetic profiles of the bacterial isolates that colonize and cause infection, thus defining if the colonizing bacteria is the same clone as that which causes the infection57,58.
Among the molecular biology methods applied in molecular epidemiology are pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST), both of which are used in studies of local and global epidemiology, respectively57.
The PFGE continues to be the “gold standard” for studies of local epidemiology, for hospital outbreaks and to evaluate transmission between hospital institutions58. It is one of the typing methods with the highest discrimination power since it allows the detection of small variations in the genome such as deletions, insertions, loss, or the acquisition of plasmids that accumulate rapidly over time58. On the other hand, the MLST analyzes the sequences of seven constitutive genes, which have a low rate of genetic variability over time57. Once the genes are sequenced, each sequence is entered into databases available on the internet in order to generate an allelic profile called a sequence type, or ST60. The relationship between STs can be determined by algorithms such as the eBURST, which allows the inference of evolutionary patterns of descent through a model of clonal expansion and diversification by assigning clonal complexes (CC)60.
Due to the availability of databases on the internet for the analysis of sequences, the MLST has facilitated a worldwide exchange and reproducibility of information60. Because of the low variability of the amplified genes, this technique is used for studies of global epidemiology in order to detect changes that are generated over long periods of time57.
Colonization and infections by multidrug-resistant bacteria in the local context
Colombia has one of the highest percentages of beta-lactam resistant bacteria, mainly Gram-negative bacilli, in Latin America61,62. The resistance to carbapenems is particularly worrisome, given that since 2006 a significant increase in this resistance has been observed in bacteria such as K. pneumoniae62. This year, the first two cases of K. pneumoniae producers of KPC-2 carbapenemase in South America were reported in a high-complexity level institution in Medellin63. In 2007, the world’s first report of this same carbapenemase was made in an isolation of P. aeruginosa that originated from the same institution61. In the following years, the situation worsened when an outbreak in 84 patients from an Intensive Care Unit (ICU) occurred in 2008, representing the first report of KPC-3 in Colombia64. Studies of the behavior of carbapenem-resistant Gram-negative bacilli in hospitals in Medellin have reported the successful international dissemination of clones, in addition to the emergence of genetically diverse clones with the capacity to harbor different mechanisms of resistance as a result of the high antibiotic pressure and the horizontal transfer capacity of mobile genetic elements61,62,65-70.
On the other hand, S. aureus continues to be one of the main bacteria isolated both in intensive care units and in other hospitalization rooms71. Previous studies conducted by our research group have shown that the strains of MRSA associated with the community have a greater capacity for virulence and dissemination than those associated with health care, and that they circulate predominantly in hospitals, displacing the clones hospital characteristics67-69.
However, most of these studies have focused on infections caused by multidrug-resistant bacteria in hospitalized patients, but very few have explored the behavior of colonization and such infections in patients on hemodialysis. A study conducted in Bogotá with hospitalized hemodialysis patients reported a percentage of colonization by S. aureus of 45.5%, with 13.3% being resistant to methicillin (MRSA)72. This leads to the need to conduct studies that provide a more complete picture of the current situation of antibiotic resistance in the hemodialysis population, patients who circulate frequently in the community and hospital institutions and who present not only an increased risk of infection, but also a greater potential for the dissemination of these resistant microorganisms inside and outside the renal unit, due to the high percentages of colonization.
Conclusion
Patients on hemodialysis are at a high risk of infection by beta-lactam antibiotics resistant bacteria and colonization could be an important risk factor for the development of such infections. It is important to know the problem of resistance in patients with a high risk of infection, not only to avoid contact of patients with these microorganisms, but to justify the need to evaluate prophylactic therapies such as decolonization.
Similarly, it is important to know the potential of dissemination of these microorganisms inside and outside of renal units, in order to establish and strengthen preventive measures to control the transmission of infections in patients on hemodialysis; a population little studied in the local area in which different risk factors converge. This knowledge will allow a first approximation of the dynamics of the transmission of these microorganisms in this population and in a country endemic to the circulation of resistant bacteria.














