Introduction
Acinetobacter baumannii (A. baumannii) a non-glucose fermenting Gram-negative bacillus, is an important cause of nosocomial infections and widely isolated from healthcare facilities, especially in the intensive care units (ICUs). They are increasingly being associated with increased morbidity and mortality among hospitalized patients. The increased use of invasive procedures and exposure to antibiotics increases the risk of infections among immunocompromised patients admitted in critical care units. These organisms are known to colonize various surfaces of the environment and patients admitted to healthcare institutions. As they are known to survive in unfavorable environments and can contaminate various surfaces present in the health care units1,2. The ability to adapt and survive in an unfavorable environment, potential to acquire multiple genes for antibiotic resistance and to colonize the patient has favored the organism to be transmitted easily and cause prolonged and repeated outbreaks3,4.
Carbapenems were considered the most powerful antibiotics because of their extremely effective antibacterial activity and low toxicity; however, with the emergence of carbapenem resistance, A. baumannii has become a growing therapeutic concern worldwide5,6. Over the last 15 years, the resistance of A. baumannii to expanded-spectrum cephalosporins and carbapenems has grown rapidly7. Carbapenem resistance, among these pathogens, is attributed to reduced expresión of outer membrane proteins (29 kDa, 33-36 kDa), change of the penicillin-binding proteins, the specific drug efflux and carbapenemase production. Carbapenem-hydrolyzing Class D oxacillinases (blaOXA-23 to 27, 40, 51, and 64 to 71) are the predominant carbapenemase has been reported globally 8,9. The blaOXA-51 is thought to be probably intrinsic to the genetic makeup of the organism. However, reports on the blaIMP or bla VIM class Metallo-β-lactamase (MBL such as blaIMP-1, 2, 4 and 5, and blaVIM-1 and blaVIM-2,) producing A. baumannii. are also on the rise 10. Although most blaNDM genes are frequently reported in Enterobacteriaceae, they have also been detected in Acinetobacter species, first of such case of blaNDM in Acinetobacter was reported in 2010 from India11. Expression of more than one type of carbapenemases has been documented in India12,13. Due to the ability to survive in a harsh environment (presence of disinfectants, exposure to antibiotics), these organisms are known to persist and are able to be transferred to different healthcare facilities by colonizing or infecting patients. Prevention of infections by these organisms is a tough challenge. Outbreaks and persistent presence of these organisms have been documented in intensive care units for prolonged periods13. The antibiogram patterns may not be able to suggest similarity among the different strains in circulation during a particular period.
A. baumannii strains have been typed using molecular methods such as ribotyping, amplified fragment length polymorphism, restriction fragment length polymorphism, and repetitive extragenic palindromic sequence-based polymerase chain reaction (Rep-PCR)14. Due to limited gene set or technical limitations, most new tests are not suitable for clinical routine monitoring in low prevalence settings. Rep-PCR is simple, rapid test that can be highly discriminatory for the identification and differentiation of A. baumannii15
The study was aimed to describe the presence of carbapenemases coding genes among Meropenem/Imipenem resistant A.baumannii obtained from clinical samples by phenotypic methods and to understand their clonal relationship using Rep-PCR.
Materials and Methods
A total of 89 non-duplicate isolates of meropenem/ Imipenem resistant A.baumannii were collected. These organisms were isolated from the samples received from patients in the critical care units (intensive care, burns, post-operative areas) of St. John’s Medical College Hospital, Bengaluru, India between May 2011 and January 2012. The isolates were collected from blood, respiratory secretions sputum, urine, pleural fluid, ascitic fluid,and wounds, and were considered nosocomial if the specimens had been obtained more than 48 hours after the time of patient’s admission. More than one isolate from the same patient was not included. A. baumannii was identified in the clinical microbiology laboratory by standard biochemical tests as well as the Vitek 2 system (bioMérieux, Durham, NC), when necessary. Those identified A. baumannii complex were further confirmed A. baumannii using a multiplex PCR method by using the specific Acinetobacter baumannii primers (P-Ab-ITSF and P-Ab-ITSB) and the internal control primers (P-rA1and P-rA2) specific for the recA gene of all Acinetobacter spp 16.The study was approved by the Institutional Ethical Board, St. John’s Medical College, Bengaluru.
Susceptibility testing
Antibiotic susceptibility was determined by the standard agar disc diffusion method using Mueller-Hinton agar plates (Himedia, India) using antimicrobial agents and interpreted according to the guidelines of the Clinical and Laboratory Standards Institute 2012 (CLSI, 2012)17. The following antibiotics Gentamicin (Gen), tetracycline (Tetra), trimethoprimsulphamethoxazole (Cotri), cephalexin (Ceph), ciprofloxacin (CFX), amikacin (Amik), Ceftriaxone and Tazobactum (Omna), cefotaxime, netilmicin (Net), ceftazidime (Fort), cefaperazone (CpZ), piperacillin (Pip); piperacillin/tazobactam (PT), Meropenem and imipenem (Imi/Mero) were tested using discs obtained from Himedia labs, Mumbai, India. Meropenem and imipenem were obtained from BD India Ltd. Resistance to Meropenem was confirmed using agar dilution method. A minimum inhibitory concentration (MIC) of ≥16 μg/mL for imipenem and meropenem was used to define resistance. Pseudomonas aeruginosa ATCC 27853 was used as reference strains for quality control of the antibiotic discs and agar dilution.
Isolation of bacterial DNA
The bacterial genomic DNA was prepared by using the lysis buffer (1% Triton X-100, 10 mM Tris pH 8,0, 1 mM EDTA) and boiling method. Briefly, colonies were picked from a MacConkey plate and inoculated into Luria-Bertani broth (HiMedia, Mumbai, India) and incubated at 37 ˚C in a shaker incubator for 18 - 24 hrs. After the incubation, cell pellet was prepared from the broth. The cell pellet was washed in molecular grade water and lysis buffer was added. The bacterial suspension was heated at 95 ˚C for 15 min, and then pelleted by centrifugation at 12,000 ×g for 5 min. The supernatant was transferred to fresh sterile 1,5 ml tubes, for use as templates for PCR amplification18.
PCR analysis of carbapenemases-encoding genes
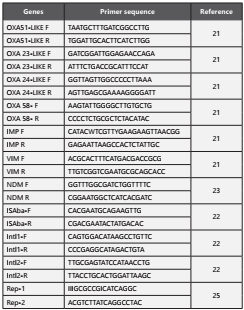
The genes detected included the integrase gene intI, OXA-type carbapenemase gene clusters blaOXA51-like, blaOXA-23-like, blaOXA-24-like and blaOXA-58-like, MBL genes blaNDM, blaIMP and blaVIM19-21. The negative control was run with every PCR. The primers used for the PCR are listed in Table 1 22 and PCR conditions are per the references quoted. The PCR products were separated on 2% agarose gels by electrophoresis, stained with ethidium bromide at a final concentration of 0.5 µg/ml, and then visualised under the gel documentation system (Biorad Life Science, CA, USA).
Rep-PCR fingerprinting
The repetitive extragenic palindromic polymerase chain reaction (Rep-PCR) fingerprinting was performed as previously described primer pair Rep-1 (5ʹ-IIIGCGCCGICATCAGGC-3ʹ) and Rep-2 (5ʹ -ACGTCTTATCAGGCCTAC-3ʹ)23,24 and are listed in Table-1. PCR products were analysed by electrophoresis on a 1,5% agarose gel and stained with ethidium bromide (5 mg/L). Strains were considered to be highly similar and belonging to the same DNA group when all visible bands of two isolates had similar migration distance. Within the same Rep-type, strains differing by no more than two bands were considered subtypes. Gel images were subsequently analysed by Bionumerics software (Applied Maths, Kortrijk, Belgium) A dendrogram was generated to analyse the similarity between the isolates, using the unweighted pair group with arithmetic averages (UPGMA) and the Dice similarity coefficient with 1% of tolerance and 0.5% of optimization25.
Results
Bacterial isolates and antimicrobial susceptibility pattern
The isolates were predominantly from respiratory secretions (endotracheal aspirates or sputum) 60 (67.4%), followed by pus 25(28.1%), urine 10(11.2%) and three from other body fluids and two from blood samples. Forty-nine isolates (55%) were from male patients. As seen in Figure-1, most of the isolates were resistant to nearly all antibiotics tested except for netilmicin. Resistance to at least one of aminoglycoside or β-lactam antibiotics were observed in 80% and 97% of the strains. A total of 36 (40%) isolates out 89 are resistant to all the antibiotics tested and rest of the 53 isolates were multidrug resistant. Minimum inhibitory concentrations (MIC) for meropenem and imipenem determined using the agar dilution method (CLSI 2010).
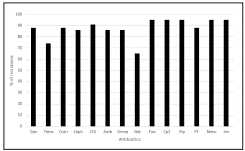
Figure 1. In- vitro antibiotic susceptibility profile of Acinetobacter baumannii species clinical isolates Note: Antibiotics tested is Gentamicin (Gen), tetracycline (Tetra), trimethoprimsulphamethoxazole (Cotri), cephalexin (Ceph), ciprofloxacin (CFX), amikacin (Amik), Ceftriaxone and Tazobactum (Omna), cefotaxime, netilmicin (Net), ceftazidime (Fort), cefaperazone (CpZ), piperacillin (Pip); piperacillin/tazobactam (PT), Meropenem and imipenem (Imi/Mero).
Detection and characterization of antibiotic resistance coding genes
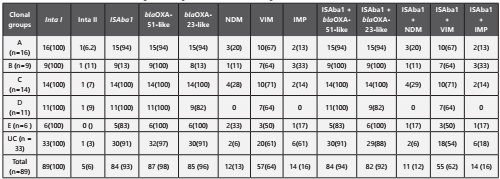
As seen in Table - 2, the majority of carbapenem-resistant isolates carried bla OXA-51(98%) and blaOXA-23 (96%) genes followed by bla VIM gene (67%). Class-1 integrase genes were found in all of A. baumannii strains tested (n=89/89,100%) whereas class 2 integrase genes were found in 5 (n=5/89, 6%) isolates. A total of 14 isolates out of 89 carries bla NDM-1 gene. The association of ISAba-1 along with blaOXA-23-likeand blaOXA-51-like was present in 92% and 94% respectively.
Rep-PCR
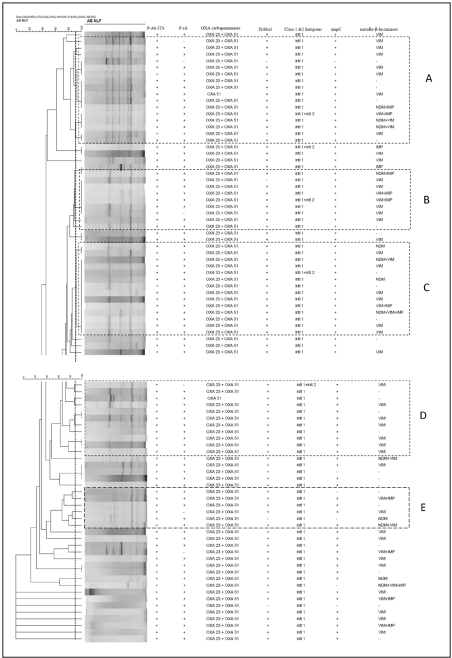
Rep-PCR showed considerable diversity of genotypes among the isolates examined. The isolates were grouped into five different clusters A to E. However, there are many isolates which has not shown any bands or showed one or two bands categorized as unclassified (UC). The clusters A to C was defined by using a similarity threshold of 90% were marked in the dendrogram (Figure 2). Majority of the isolates from all the clusters demonstrated the presence of blaOXA-23, blaOXA-51 and class-1 integron coding genes. Similarly, isolates in Cluster B carried blaVIM, in few isolates, blaVIM coding gene coexisted with blaIMP and one isolate carried both bla NDM and blaIMP coding genes. The Clusters D to E was defined by using a similarity threshold of 90% as marked in the dendrogram. Isolates in cluster D in addition to bla OXA-23, bla OXA-51and class-1 integrons coding genes, demonstrated only the presence of blaVIM gene. The isolates grouped under Cluster E carried genes code for blaVIM, blaIMP and blaNDM.
Discussion
Reports on carbapenemase - producing A. baumannii isolates are rising globally due to increased carbapenem use thereby creating a selection pressure. In the current study, we found that most of the carbapenem-resistant isolates (67%) were from respiratory secretions. Outbreaks caused by A. baumannii strains have been reported in the ICU due to cross-contamination or spread among patients1,26,27. It has been reported that carbapenem resistance rates in A. baumannii have mostly exceeded 40% throughout all of India. All of the isolates tested in the present study showed a high level of resistance to majority of the antibiotics except netilmicin. Prashanth et al., report the high effectiveness of netilmicin against MDR A. baumannii from India28. However, Saranathan et al., also reported 48% of netilmicin resistance among A. baumannii clinical isolates from Southern India29. Infections like bacteremia and ventilator-associated pneumonia caused by XDR A. baumannii result in >50% mortality rates30. In the present study, 80% of A. baumannii isolates were detected as XDR. Rynga et al., report 78% XDR A. baumannii clinical isolates from two hospitals from New Delhi, India31. Similarly, Maspi et al., report 71% of XDR A. baumannii isolates from Iran32. Multi-drug resistant A. baumannii has caused many nosocomial outbreaks worldwide, and is considered as a difficult to treat nosocomial pathogen due to resistance to many antimicrobial agents and has been increasingly associated with mortality10,13.
The higher percentage of oxacillinases, blaOXA-23andblaOXA-51 (most common resistance genes), than metallo-β-lactamases (blaVIM and blaNDM) detected from the tested A. baumannii isolates was in accordance with earlier published reports from India12,33. However, in the present study, blaOXA- 58-like carbapenem resistance coding genes was not detected from any of the tested isolates. The blaOXA-58 like genes have been more commonly reported from Europe, North and South America, and West Asia, however low prevalence (2%) has been reported from South India34. In this study we observed, both blaOXA-51 gene and blaOXA-23, were important in conferring resistance to carbapenems among 91% of the isolates which was comparatively higher to other Indian studies34. In our study, approximately 93% of the imipenem/carbapenem-resistant isolates carried the combination of blaOXA-51and blaOXA-23 along with insertion sequences ISAba1. Vijayakumar et al., reported the association of ISAba1 and blaOXA-23 gene among Carbapenem resistantA. baumannii from a Tertiary Care Hospital in South India35. However, Ganjo et al., report the blaOXA-23 gene in combination with ISAba1 insertion element was largely responsible for carbapenem resistance in clinical A. baumannii isolates from Kurdistan, Iraq36. Present study report 11/12 blaNDM and blaOXA-23 coding gene carried isolates associated with ISAba1. Karthikeyan et al., reported the co-existence of blaNDM gene and blaOXA-23 with ISAba1 from India11. A total of 14/89 isolates carried blaIMP coding gene, out of these 14 isolates, four isolates carried blaNDM and blaOXA-23 associated with ISAba1. However, the coexistence of blaNDM-1 with the blaOXA-23 and blaIMP among A. baumannii isolates was reported from China37. Carbapenem resistance in Europe is mostly due to the occurrence of blaOXA carbapenemases, however, the blaIMP and blaVIM MBLs are more prevalent in the Far East. Present study detects blaVIM Metallo-β-lactamses coding genes from 64% of A. baumannii isolates, among these 7/89 (8%) co-existed with blaNDM. Rhamanet al., reported blaVIM was significantly associated with bla NDM genes in A. baumannii isolates from India38.
Incretion Sequence elements have significant role in A. baumannii antimicrobial resistance. ISAba1, ISAba2, ISAba3, ISAba4 and IS18 are frequently associated with the expresión of carbapenemases genes in A. baumannii. The present study observed a prevalence of 94% and 92% of ISAba1 elements associated with bla OXA-51-like and blaOXA-23-like genes respectively. Similarly, Villalón et al. (2013) investigated the presence of IS elements in 59 multidrug-resistant A. baumannii isolates and observed a prevalence of 93.2% of ISAba1 from Spain.
It is important to note that IS elements such as ISAba1 can contribute to the spread of carbapenemase genes among different Acinetobacter species39. The high prevalence of class 1 integrons among multidrug-resistant A. baumannii clinical isolates has been confirmed worldwide. The epidemic potential of A. baumannii may be linked to the presence of class-1 integrons and ISAba1. In our study, class-1integrons was present in all of A. baumannii isolates. Whereas, class 2 integrons were present in four isolates and co-existed with class -1 integron. In the present study, 93% of A. baumannii isolates carried class-1 integrons coding genes and these isolates co-existed with ISAba1. Sun et al report 64% of the class -1 integrons in Acinetobacter spp. isolates in northeastern China40. Amin et al reportedthe class -1 integron is often responsible for the dissemination of the MBL genes among A. baumannii isolates from Iran41. In the present study, 83% (74/89) of A. baumannii isolates were positive for class-1 integrons coding genes.
This study was also aimed to determine the distribution of resistant genes in genetically similar and dissimilar carbapenem-resistant A. baumannii strains by using Rep-PCR. All the isolates were classified into different clonal clusters A to E according to genetic similarity based on banding pattern obtained by Rep-PCR. Resistance coding genes detected from these isolates were not associated with a particular group; rather each group confined more than one resistance gene type. Similar to other study, most of the strains showed to harbor multiple carbapenemases genes possibly indicate that there are multiple clones in circulation in our hospital. In this study, it has been shown that genetic determinants like blaOXA- 23,blaOXA-51, intI1, ISAba1, blaVIM., blaNDM,andblaIMP were widely distributed among all the carbapenem resistant A. baumannii isolates grouped under different clonal clusters A to E. These genetic determinants are capable of possessing the potential horizontal gene transfer between susceptible A. baumannii or any other bacterial species. Rep-PCR uses amplification of intervening fragments between highly conserved sequences by means of consensus primers14,15. It is a rapid, simple and lowcost method for molecular typing, which has been applied previously to identify genetic differences among the isolates of A. baumannii15. It is challenging to discriminate a clear pattern among some isolates because rep-PCR bands are faint, and some profiles have few bands. The rep-PCR is less discriminatory than PFGE in identifying unique strains. However, sometime it requires isolates sharing the same pattern to be tested with a more discriminatory method, like PFGE or multilocus sequence typing whenever appropriate. Vijayakumar et al reported the carbapenem-resistant A. baumannii isolates were grouped under clonal complex 208 which belongs to the international clonal lineage 2 and high occurrence of ST-848 carrying blaOXA-23-like gene from India42.
There has been increasing concern regarding the rise of Acinetobacter infections in critically ill patients. Our study also showed that Acinetobacter infections most frequently involve the respiratory tract of intubated patients as also reported by others43. The ICU and other critical care áreas provide an ideal environment for the persistence of this organism in the presence of intense antibiotic exposure, enhancing mutation or acquisition of additional resistance mechanisms. The ability of this organism to acquire resistance from other gramnegative bacilli has been demonstrated by the presence of the blaNDM gene, first reported from Enterobacteriaceae3. Multiple hospital admissions and visit to different institutions increases the risk of colonization with different strains of the pathogen33. Despite appropriate infection control precautions these organisms have been reported to cause prolonged outbreaks among patients admitted in the ICUs26.
Conclusion
Abaumannii previously thought to be contaminants have now gained notoriety as multidrug resistant, hospitalacquired pathogens due to the acquisition of a number of resistance genes. Analysis of the genes that codes for carbapenem resistance among randomly selected isolates has confirmed the presence of multiple resistant genes such as, bla OXA-23, blaOXA-51, and blaVIM and isolates that showed considerable genetic heterogeneity. Presence of blaNDM was documented along with other Metallo-β-lactamases genes. In most of isolates the combination of genes demonstrates the ability of the organism to acquire novel genes from other bacteria (such as blaNDM) and increase its own repertoire of resistant genes, thereby markedly reducing treatment options with β-lactam antibiotics.