Introduction
Throughout the history of mankind, infectious diseases have been one of the causes of higher mortality. Thanks to the discovery of antibiotics, one of the most important findings in the evolution of clinical medicine, those figures decreased significantly. However, the ability of microorganisms to mutate when exposed to antimicrobials has led to the inefficiency of treatments and the excessively over- and misuse of antibiotics intensifies the development and dissemination of drug resistant bacteria.
According to the World Health Organization (WHO) antibiotic resistance poses one of the major challenges to global health. Currently, medical personnel face uncertainty when making therapeutic decisions, since many common pathogens such as those causing tuberculosis, pneumonia, or infections of the digestive and urinary tracts, which were previously easily eliminated with antibiotics, have developed drug resistant mechanisms that hinder their treatments.
The pediatric population is directly affected by this problema due to the high incidence of infections in children and its associated use of antibiotics. An intercontinental study, which evaluated 226 hospitals from 41 countries, shows that 36.7% of the pediatric patients received at least one antimicrobial in 2012, being antibiotics the most commonly used. That percentage was increased to 61% in pediatric intensive care units1.
One of the few studies about antibiotic resistance in dominican pediatric patients sought to identify the resistance profiles of Streptococcus pneumoniae against non betalactam antibiotics in 10 Latin American countries. In that study, among the 415 isolates from Dominican Republic (DR), 12.3% were resistant to erythromycin 1.4% to chloramphenicol, and approximately 53.3% to trimethoprim sulfamethoxazole2.
The significant rise in the prevalence of infections caused by resistant microorganisms is one of the greatest threats to public health systems in our country, as well as globally, since these infections are associated with increased cost in medical care, longer hospital stays and higher morbidity and mortality rates. Due to the lack of reliable information in DR about the extent of the problem in pediatric services, our research team decided to evaluate the resistance profiles of pathogenic bacteria isolated from third level centers over a period of two years.
Methods
We conducted a descriptive, retrospective study on reports showing positive bacterial cultures from patients admitted in the pediatric unit of three third level hospitals in the city of Santiago de los Caballeros, DR, from January 2016 to December 2017. Those three hospitals were the only medical centers with a data base of microbiological reports in the Cibao Region. This research was approved by the Faculty of Health Sciences Bioethics Committee (COBEFACS).
The different hospitals performed the sample collection and bacterial cultures following their approved and standardized protocols. Bacterial identification and antibiotic susceptibility testing were performed using automated systems, two of the hospitals used Vitek 2® and one MicroScan® systems. We defined four antibiotic resistance profiles: isolates resistant to at least one extended spectrum cephalosporin (cefotaxime, ceftazidime, and/or ceftriaxone) were labelled as third generation cephalosporins resistant (3GCR); carbapenem resistant Enterobacteriaceae (CRE) were identified as resistant to at least one carbapenem drug (ertapenem, imipenem, and/or meropenem)3; regarding Staphylococci, methicillin resistance was determined if isolates were resistant to oxacillin; inducible clindamycin resistance was established when the microorganism was erythromycin resistant and clindamycin susceptible. Confirmatory tests for clindamycin resistance are not commonly performed in the local clinical labs, thus, most of the isolates identified as inducible clindamycin resistant were reported so based only on the automated system results. The interpretation of the antibiogram results was done according to Clinical and Laboratory Standards Institute (CLSI) guidelines.
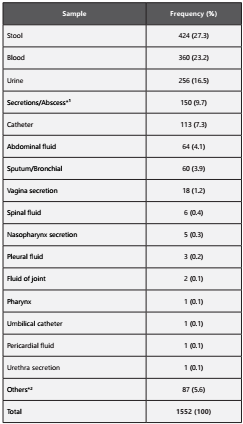
We obtained an initial sample of 1884 culture positive reports. 332 were excluded because they reported more tan one isolate or not all the necessary antibiotics to define the phenotypes were tested, resulting in a total of 1552. Also, we were unable to include reports identifying Enterococcus spp. because its antimicrobial susceptibility was not tested in the different hospitals; and, among the Gram positive isolates, only those identified as Staphylococcus spp. were analysed, as only one of the three centers tests Streptococcus spp. samples, in both cases probably due to technical complexity. We included clinical specimens from all the possible sources: blood, urine, stool, catheters, secretions and others (Table 2).
We collected data from cultures and antimicrobial susceptibility results from the clinical laboratories’ database and we entered it in the WHONET software for the Surveillance of Antimicrobial Susceptibility (https://www.who.int/medicines/areas/rational_use/AMR_WHONET_SOFTWARE/en/). We exported the generated file to Microsoft Office Excel, where the data was cleaned, eliminating all duplicate or incomplete reports.
Results
During our study, we analysed a total of 1552 positive culture results from which 1041 were Gram negative rods and 511 Gram positive cocci. The most commonly identified microorganisms are shown in Figure 1. Of the 1041 cultures reporting Gram negative microorganisms, 524 (50.3%) were described as 3GCR and 179 (17.2%) as CRE. Additionally, 129 (12.4%) isolates exhibited both phenotypes (Table 1.A). Remarkably, Serratia marcescens was revealed as our second most frequent Gram negative microorganism, and almost all of those isolates (117 out of 122) were 3GCR (95.9%), while the frequency between our other species ranged from 30 to 70% (Table 1.A). On the other hand, only one of the Serratia isolates was CRE (0.8%), while the other species ranged from 5 to 48%.
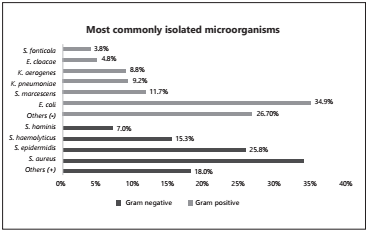
Figure 1 Percentage of the ten most frequently identified microorganisms. The Gram negative species more frequently reported in this study are represented in the upper half of the figure, which include a total of 1041 reports (100%). The Gram positive pathogens are in the bottom half, with 511 reports (100%). Gram negative: Escherichia coli (E. coli), Serratia marcescens (S. marcescens), Klebsiella pneumoniae (K. pneumoniae), Klebsiella aerogenes (K. aerogenes), Enterobacter cloacae (E. cloacae) and Serratia fonticola (S. fonticola). All the Gram positive species belong to the Staphylococcus genus.
Table 1: Frequency of 3GCR and CRE in Gram negative microorganisms (A), and methicillin and inducible clindamycin resistance in Gram positive microorganisms (B), classified by species.
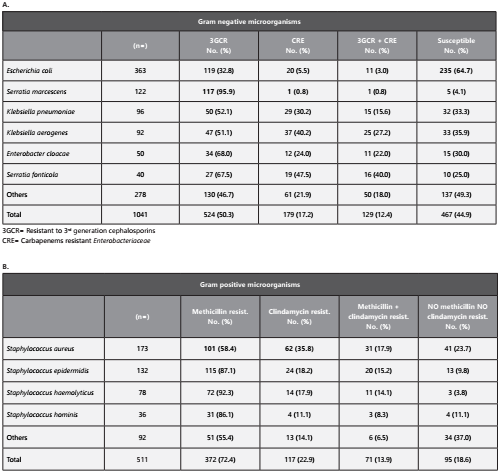
Furthermore, among the 511 cultures identified as Gram positive cocci, 101 (58.4%) of the 173 S. aureus isolates were resistant to methicillin (Table 1.B). Also, 269 (79.6%) out of 338 of the coagulase negative Staphylococcus, which are frequently found colonizing the skin of patients and hospital employees, showed resistance to methicillin (Table 1.B). We also found that 117 (22.9%) of the Gram positive isolates showed inducible clindamycin resistance, with S. aureus exhibiting the higher rate (35.8%), and a total of 71 (13.8%) isolates presented both phenotypes (methicillin and inducible clindamycin resistance) (Table 1.B).
The microorganisms included in our study were isolated from different sources: stool 426 (26.5%), blood 375 (23.3%), urine 270 (16.8%), catheter 117 (7.3 %), secretions 150 (9.7%) and others (Table 2). Among the blood samples, the most frequently isolated microorganisms were S. marcescens (26.1%), Staphylococcus epidermidis (13.3%), Klebsiella pneumoniae (11.7%) and Staphylococcus haemolyticus (6.4%). On the other hand, 47.4% of the 270 isolates obtained from urine samples corresponded to E. coli, which exhibited moderately high sensitivity (64.7%, Table 1.A)
Discussion
We report here the most commonly identified pathogens found in pediatric services in the Dominican Republic (Figure 1), which match most of the similar previously published reports, where the most frequently described microorganisms are almost always members of the Enterobacteriaceae family, Staphylococcus aureus, and coagulase negative Staphylococcus4. Among the Gram negative microorganisms analysed, we found similar resistant patterns to those reported by Legese et al. from Ethiopian children, where 22 out of 28 (79%) of Enterobacteriaceae isolates were ESBL producers and 12% carbapenemase producers5; and by Apondi et al (2016) in Kenya, where 23% of the samples from children and newborns were identified as K. pneumoniae, of which more than 80% were ESBL and 23% were CRE6. By contrast, the prevalence reported by recent studies for beta-lactam resistant S. marcescens is normally much lower than the one we report, as it is usually close to 50%7. The number of Serratia isolates reported was also strikingly elevated, but at this moment we cannot ascertain if those resistant isolates derived from a specific outbreak or it was due to a regional or local characteristic.
Surprisingly, we found that 4.8% of the analysed isolates presented resistance to at least one carbapenem drug, but not to any of the three extended spectrum cephalosporins studied. Since this study is retrospective, we cannot know the specific resistant mechanism used by those isolates.
Our results on the Gram positive cocci are also congruent with previous reports from other Latin American countries4. These data are worrisome, as patients suffering from methicillin resistant S. aureus (MRSA) infections need greater use of antibiotics, longer hospital stays and more diagnostic tests8.
When we compared the main pathogens we found in blood samples with one of the most recent studies on pathogens isolated from blood infections in children, conducted in the hospital Dr. Hasan Sadikin in Bandung Indonesia, we found that, in both cases, the most common bacteria causing these infections were also Enterobacteriaceae and staphylococci, although the specific species varied9.
Similarly, E. coli was reported as the most prevalent uropathogen, which is not surprising10. Furthermore, E. coli presented 33% of resistance to third generation cephalosporins in our study, which is similar to other Latin American countries11, but still alarming. The application of empirical antibiotic therapy with broad spectrum antibiotics is regularly used to treat urinary tract infection, which are very common and severe in children. Therefore, E. coli usually shows alarmingly high resistance to commonly prescribed antibiotics in the pediatric population, especially in countries where antibiotics are available over the counter10. It is also usual to find in different studies a higher resistance to betalactam antibiotics than to carbapenems, with a similar pattern to our current study (Table 1.A)12
The main limitations of our study are 1) we cannot differentiate between colonisation or infection; 2) the lack of confirmatory tests, such as the D test for clindamycin resistance; and 3) the lack of phenotypic and molecular characterization of the CRE and cephalosporin resistant strains. Thus, some clonality could be possible, for example in the Serratia isolates. Also, only four phenotypes were analysed to determine the resistance profiles presented by the microorganisms. In a future study, it would be useful to analyse the resistance to specific antibiotics, allowing a more complete evaluation, including, for example, resistance to cefoxitin to detect AmpC producing microorganisms. Moreover, future studies should also focus in the molecular characterization of the specific genes that are causing the resistant phenotypes in order to tackle antimicrobial resistance in the country, which is alarmingly elevated according to our present study. This data could help to avoid the use of potentially toxic drugs.
Nevertheless, our investigation has several strengths, since it is a multicentric study and includes a significant sample size and evaluation period. But most importantly, this study entails the first descriptive report of resistant patterns, expressed by bacterial pathogens, affecting pediatric patients in the DR. In consequence, the data found in our research would allow the practitioner to provide more effective empirical antibiotic therapy to those individuals that require immediate treatment, taking into account the most frequent pathogens and their resistance profiles. For example, based on our results, infections suspected or confirmed to be caused by S. aureus should include an anti MRSA drug, and those suspected or confirmed to be caused by S. marcescens should not be treated with third generation cephalosporins if antimicrobial susceptibility test results are not available. We also expect that the information collected in this study will créate awareness in the general community, the medical population, and those responsible for creating laws or legislation at both public and private levels.