Introduction
Worldwide, the diagnosis of HPV-associated cancer is increasing2. Despite current prophylactic interventions, a significant proportion of patients suffers a cancer-specific mortality3, which leads to a global awareness of the importance of identifying factors associated in the etiology of cancer. The latter could provide a better understanding of preventive, predictive and public health purposes.
According to this, HPV-DNA integration into human genome is frequently an important event in the pathogenesis of HPV-associated cancer4. The virus-mediated carcinogénesis has been mediated by two important pathways: Deregulation of viral gene expression and genomic instability of the host5. Accordingly, there is experiments describing that integration could result in augmented levels of oncogene (E6/E7) transcripts6. The selective growth and the genomic instability of the cell come from the integration process7. Furthermore, host genomes with integration of HPV have also different expression and methylation profiles compared with nonintegrated genomes8.
HPV integration is considered a process which occurs randomly in almost all chromosomes9. However, some important regions of the genome have been found to have repeatedly integration of HPV10. Other genetics elements surrounding the integration site are involved in the integration evento and enhance the genomic instability. These elements are part of genomic environment, for example, it has been indicated that HPV integration occurred within - or neighboring - sequences of Alu-repeats11. Additionally, due to persistent infection of HPV, it becomes susceptible to epigenetic modification (CpG Islands and rich CG regions)12. Another element is DNase I hypersensitive sites where studies provide evidence that DNase I sites are surrounding the host-viral junction13,14. Finally, identifying genes with HPV integration and evaluating subsequent alterations in the expression, turn the transcriptional regions into important elements to analyze. However, the intergenic sites must be taken into account because viral integration into these frequently occurs in cervical cancer and it has also been identified in oral squamous cell carcinoma15.
The aim of this study was to identify in silico, molecular regions of the genome where the HPV16 integration events occur.
Methods
We performed a bioinformatic study based on a systematic search in Medline through PubMed, Embase and Lilacs from inception to April 2019. Our search included the following terms: “((HPV) OR human papillomavirus) AND ((cancer) OR carcinoma) AND ((integration) OR breakpoint)”.
Inclusion criteria: We included studies that reported HPV integration places and focused on published articles. We did not impose any language restriction. Two reviewers selected the studies by title, abstract and full-text. The genome browser positions or nucleotide sequences adjacent to the viral integration site of the selected articles were extracted.
Exclusion criteria: Those articles showing no information regarding integration places.
In-silico methods
It consists of two components: 1) The search and retrieval of the integration sites reported for different HPV genotypes, and 2) The evaluation of the integration sites in a descriptive way (chromosomal distribution and clinical variables) and through bioinformatics tool. We used the UCSC Genome Browser Home (https://genome.ucsc.edu) to evaluate the transcriptional units, intergenic sites, DNase I hypersensitive sites (DHSs), Alu sequences and CpG islands present on viral breakpoint. The reference genome GRCh38-2013 was used for the alignment of nucleotide sequences.
A descriptive analysis of the data obtained from each of the variables was performed related to the HPV-16 genotype. Additionally, through IBM SPSS Statistics 20 a statistical analysis was carried out to identify significant differences between the content of CG dinucleotides in exons and introns with integration sites, to test for major integration events in hypomethylated regions. Finally, the genes where an HPV integration site was found were analyzed through gene set enrichment analysis. This analysis can find significant associations with functional gene sets.
Results
We found 1808 studies according to the search strategy; then we included integration sites from 20 studies. The integration sites were distributed on all chromosomes and 667 integration site were obtained (Figure 1). Chromosome 19 had the highest number of integration sites (23 integration sites).
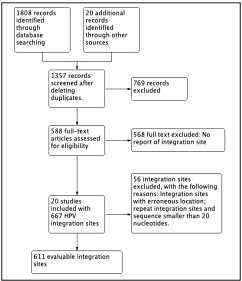
Figure 1 Flow diagram of In silico methodology. In total, 611 integration sites were analyzed for HPV-16
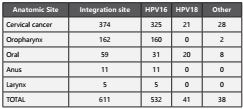
When analyzing the HPV integration sites by anatomical location, we found that 374 (61%) were related to cervical cancer. In addition, 325 (87%) of these integration sites had HPV-16, 21 (5%) had HPV-18 and 28 (7%) had another type of genotype. Oro-pharyngeal cavity was the second anatomic site with 162 (26%) integration sites. It is noteworthy that the HPV-16 was found integrated into 160 (99%) analyzed sites. The oral cavity presented 59 (10%) integration sites where HPV-18 showed a higher frequency (49%) compared to HPV-16 (31%). A remaining 20% presented other genotypes of HPV. Finally, the anatomical sites anus and larynx showed 11 (2%) and 5 (1%) integration sites respectively (Table 1).
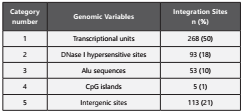
When analyzing the different genomic variables, 50% of HPV16 integrates into transcriptional units (Table 2). 306 sites, coming from this group were divided into 24 integration events in exonic regions (8%, corresponding to 19 genes) and 282 integration events in intronic regions (92%, corresponding to 163 genes).
From 182 genes present in the sites of viral integration, four presented integration events in both exonic and intronic regions, therefore, a total of 178 genes involved in viral integration events were shown. Additionally, we found that the exons tend to be more methylated which could explain the tendency of HPV to integrate into intronic regions. The percentage of CG in the exonic region was 55.91% (95%CI: 45.34-66.47) while in the intronic region was 44.03% (95%CI:41.32-46.73). The percentages of CG in the two regions were significantly different (p = 0.015).
Furthermore, three out of 178 genes, were tumor suppressor genes (TP63, PRBM1, and FHIT) and four were oncogenes (ERBB4, MECOM, MLLT1 and USP4) (Figure 2).
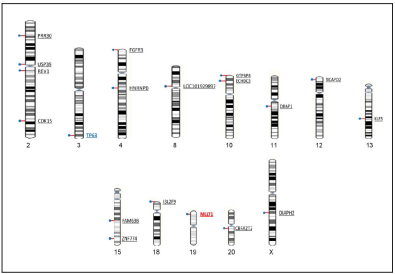
Figure 2 Viral integration sites in exonic regions. In total 21 viral integration sites were found in exonic regions of 19 transcriptional units. The human genes USP39 and TP63 showed two vira integration events. The TP63 is a suppressor gene and the MLLT1 is an oncogene.
Intergenic sites and sites of DHSs were present in approximately 20% of the integration sites; Alu sequences were present in 10% of integration sites and CG rich regions were shown in only five (1%) of the integration sites included in the study.
Discussion
In the in-silico review, 611 of 667 integration sites for the HPV 16 genotype were collected, with a high proportion of squamous cell carcinomas (91%) and 60% in cervical-uterine anatomical sites, followed by 30% in oropharyngeal anatomical sites.
Analyzing the proportion of integration sites by chromosome size, a greater proportion of integration sites were found on the small chromosomes 19, 20 and 21, followed by chromosome 9. Also, chromosomes 9, X, 20 and 21 were found to have a high frequency of affected gene regions, according to the number of total genes per chromosome. The previous results show that chromosomes 9, 20 and 21 have a posible relevance in the integration event because they present the highest proportions of integration sites according to their size and number of total genes, also, it could be suggested that that integration is not a random event such as indicate some authors9,16.
With respect to the integration sites by anatomical site, it is clear that cervical cancer presents a higher frequency of integration sites reported in the literature. Cervical carcinoma is one of the most commonly occurring cancers in women worldwide, accordingly, greater than 99% contains HPV sequences17. On the other hand, oropharyngeal cancer has also reported a large number of integration sites in the literature. HPV infection of the mouth and oropharynx can be acquired by a variety of sexual and social forms of transmission. HPV-16 accounted for a larger majority of HPV-positive oropharyngeal SCCs (86.7%) than HPV-positive oral (68.2%) and laryngeal SCCs (69.2%). HPV-18 was the second most frequent type detected: 2.8% in oropharyngeal, 34.1% in oral, and 17% in laryngeal SCCs. Other oncogenic HPVs were rarely detected in HNSCC18. Our data on HPV-18 could suggest that the frequencies reported in some scientific articles may be underestimated.
A significant result in the analysis of genomic variables was that 50% of HPV16 integration events occur in coding regions or transcriptional units. This result agrees with the found by Zhang and colleagues from the analysis of 14 cervical cancer publications where these integration sites showed preference for transcriptionally active regions and intragenic áreas19. Furthermore, a higher proportion of integration events occur in intronic regions (92%), results that are consistent with the reported by Christiansen and colleagues in 201520. A total of 178 coding genes were affected by the HPV integration event.
The 178 coding genes were subjected to a Gene Set Enhancement Analysis (GSEA) focused on their biological function. 59 of these genes were part of the following biological processes: tissue development, expression of genes of active regulation, the formation of anatomical structures, biosynthetic processes of positive regulation, regulation of transcription, cell cycle and development of epithelium. The other 119 genes each are part of biological processes from many other metabolic pathways.
In the 178 genes, three were tumor suppressor genes (FHIT, PRBM1 and TP63) and four were oncogenes (ERBB4, MECOM, MLLT1 and USP4). The importance and relevance of integration sites on the tumor behavior is still unclear nowadays, however, understanding the complex of genes and viral integration with the disturbances of cellular expression might allow changing the oncologic therapies for patients15.
Additionally, integration sites were found to coincide with DNase I hypersensitive sites in 18%, sites that have been assigned as regions available for the integration evento20 and associated with genomic instability, A characteristic of human cancers. Alu sequences were found in 9.5% of integration events, close to that reported by Zhu and colleagues (14%)21.
As for regions rich in CG dinucleotides, only one integration site contained a CpG island, possibly because DNA methylation may act as a barrier to virus integration in these regions.22 DNA methylation of the viral upstream regulatory region (URR) has been additionally associated with a latent and persistent infection23.
There has been found a correlation between the extension of the viral quiescence, the DNA methylation, and the regulation of the viral genome expression in those viruses with the ability to maintain as a latent episome24.
As a conclusion, our results suggest that many of the integration sites reported in the scientific literature are HPV 16 from squamous cell carcinomas (91%). 60% of these carcinomas belonged to the cervical anatomical sites and 30% to the oropharyngeal anatomical sites. It is also interesting to highlight that 50% of HPV16 integrates into transcriptional units and might affect the expression of target genes.