Introduction
The Hepatitis C virus (HCV) is a Flavivirus of the Hepacivirus genus, which infects principally liver cells and which can cause cirrhosis, hepatocellular carcinoma, and liver failure. It has been considered the principal cause of chronic liver disease with an annual estima te of 700,000 deaths globally with estimates of approximately 180-million people infected with the virus1,2. Between 75% and 85% of the people have chronic infections and only 21% have acute infection and, given that it is generally a subclinical disease, its initial diagnosis is complicated1,3,4. Its management is based principally on pegylated interferon α and ribavirin; however, their effectiveness is limited for genotypes 1 and 4, requiring addition of viral protease inhibitors telaprevir and boceprevir, improving response to treatment by 80% with genotype 1, but - in many cases - the treatment may not be effective.
Additionally, HCV can replicate in peripheral blood mononu clear cells, dendritic cells, and cells of the central nervous sys tem through interaction with receptors, like DC-SIGN (CD209) and L-SIGN (CD209L), heparan sulfate (CD44), lipoprotein re ceptors, scavenger receptor B1 (SR-BI), tetraspanin-28 (CD81), Niemann-Pick C1-like 1(NPC1L1), claudine-1 and occludin, epidermal growth factor receptor (EGFR), and Ephrin type A2 receptor (EphA2) through clathrin-mediated endocytosis5,7.
CD209 and CD209L receptors are important contributors to the initial establishment of the infection by HCV, enabling the virus to cross through different cell types until recognizing hepatocytes, which are the target cell for virus replication. HCV forms complexes with lipoproteins that allow the virus to gain proximity to the hepatic cellular surface through HSPG, LDLR, and SR-BI receptors to, finally, closely bind to CD81 and CLDN1 receptors. Said receptors are recognized by the envelope protein, a heterodimer formed by E1/E2 proteins, Particularly, the E1 protein has a determinant function in the fusion of membranes for release of viral nucleocapsid inside the cell. Furthermore, it helps the E2 protein in recognizing hepatocytes. Also, the E2 protein has been better characteri zed and it has been demonstrated that it is indispensable for initial recognition, distribution, and establishment of chronic infections in the liver through especially CD81. The E2/CD81 interaction generates the formation of an endosome in the cell interior, where a change in pH induces a conformational change in the envelope protein, allowing membrane fusion and release of the viral genome into the cell5,8,9.
The lack of cell models that permit in vitro assessment of compounds may explain the shortage of studies seeking the rapeutic alternatives to treat infection by this virus, hence, the search for new molecules that complement treatment and which, additionally, manage to bear in mind the impor tance of the biological host-virus interaction, are of great in terest to HCV and permit facing challenges when established treatments fail2,10-13.
One of the alternatives proposed in this work is the use of inhibitory peptides of protein interactions (iPPI), studied to inhibit interaction processes between two proteins as biolo gical target14-16. This type of peptide molecules has attracted the attention of researchers given their capacity to recognize protein interactions or specific targets with high selectivity. Additionally, these molecules show high tolerance to the organism based on their amino acid composition and are easily synthesized and modified to improve their chemical and physical stabilities and half-life in plasma. These features have allowed the development of currently FDA-approved peptides for clinical use17-19.
Several studies seeking anti-HCV peptides have used in si lico approaches, such as the study conducted by Chang et al. in 201720, who propose peptides that block the CD81 cell receptor as a mechanism to prevent binding to the E2 vi ral protein and an approach to determine the importance of this receptor in viral infection. Yin et al. 201721 proposed four peptides against E1 and E2 proteins that inhibit virus entry to the cell. Additionally, the E2 protein is studied to evaluate its potential as a vaccine target; therefore, Hua et al. 201722 assessed conserved sequences of the HCV E2 protein to pre dict B-cell epitopes. Moreover, Dawood et al. 201923, studied peptides derived from E21E2, NS4B, NS5A, and NS5B that were synthesized as multiple antigenic peptides to stimulate and produce a humoral immune response.
All of these studies rely on complex bioinformatics tools and computational servers to determine the molecular structures and analyze the predicted interactions using mathematical models. Computational tools do not determine the effective use of different molecules; however, these methods provide important theoretical approximations to predict aspects such as molecular interactions and behavior, structure, side effects, among others. In silico approaches enable an initial screening of candidate compounds prior to their evaluation in vitro and in vivo; thus, reducing the costs of this type of studies24,25.
Given the aforementioned, this work sought to design, via computational methods, inhibitory peptides of protein inte raction between the E2 protein of the HCV and the CD81 and CD209 cell receptors by using open-access web servers.
Methodology
Selection of protein structures to generate interaction complexes
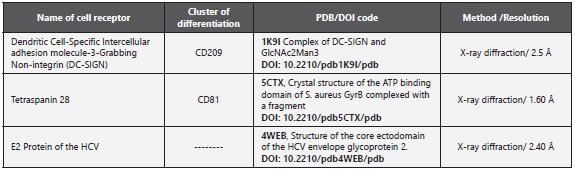
Protein structures crystalized through high-resolution <4Å X-ray diffraction were selected from the Protein Data Bank database (https://www.rcsb.org); the receptors elected were the Human CD81 and CD209 given the importance of these receptors in cell recognition by the virus; furthermore, the vi ral ligand was the E2-HCV protein, given its direct interaction with diverse receptors (Table 1).
E2-HCV protein sequence analysis and variability among viral genotypes
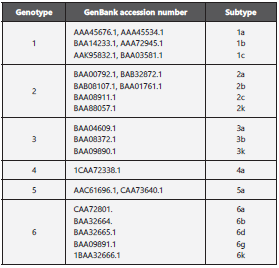
The variability of E2-HCV among genotypes 1-6 of the he patitis C virus was evaluated through multiple alignments using ClustalOmega (https://www.ebi.ac.uk/Tools/msa/clus talo/). The complete polyprotein sequences were retrieved from GenBank (Table 2) and aligned to the E2-HCV protein sequence (PDB: 4WEB) used to design the peptides.
Design of interaction complexes between the E2 protein of the HCV (E2-HCV) and the CD81 and CD209 cell receptors
Upon selecting the crystals from the receptors (CD81 and CD209) and the E2-HCV protein, an interaction complex was carried out between CD81/E2-HCV, CD209/E2-HCV through the ClusPro 2.0 web server (https://ClusPro.bu.edu/help.php) from Boston University. According to Kozakov et al., 201726, this server performs three computational steps; a rigid cou pling based on billions of conformations, a grouping based on the root mean square deviation (RMSD) of 1000 structu res with the lowest energies generated to find the biggest groupings represented by the most-probable models of the complex, and, finally, refinement of selected structures by using energy minimization to obtain the ten best models. From these groups, the server generates four sets of models: balanced, electrostatically favored, hydrophobically favored, and van der Waals + electrostatic24,26-29. To conduct the inte raction complexes, the E2-HCV protein was used as ligand, (PDB: 4WEB) and as receptors, the CD81 (PDB:5CTX) and CD209 (PDB:1K9I). From the results obtained, the balanced models were selected, without favoring any of the forces that could dominate over the model.
In silico design of peptides derived from the protein-protein interaction (iPPI)
Upon creating the CD81/E2-HCV and CD209/E2-HCV inte raction complexes between the cell receptors and the E2- HCV, the study used the Peptiderive tool from the Rosetta Online server (https://rosie.rosettacommons.org/peptideri ve/)25,30,31; which is an open-access program that generates linear and cyclical (suggested) peptides that recognize key segments (hotspot) with significantly low energy bonds that interfere in the interaction of the complex of two proteins that have been modelled.
Peptides were designed with lengths of 18-20 amino acids and were considered as candidates peptides with interface scores (Rosetta Energy Unit, REU) below -10, given that be low this value, the peptide’s contribution to the total binding energy of the complex is between 35% and 50% of relative energy and at values above -10, the contribution percenta ge is lower. The REU is a scale of interaction not equivalent to the measurement of the amount of energy expended per number of molecules (kcal/mol); it is server-specific and is on an arbitrary scale, and although it does not represent the lowest energy values, it does use a combination of different energy functions25,30.
Obtaining the theoretical physicochemical and cytotoxic properties
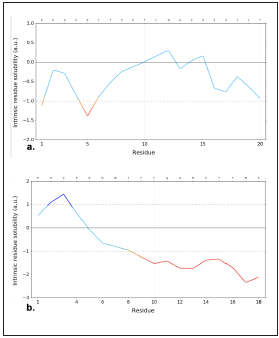
Given that we expect to have in the future safe peptide mo lecules to use in living organisms, an approach was made of their theoretical physicochemical behavior (in silico) through the ProtParam platform https://web.expasy.org/protparam/, Pepcalc platform https://pepcalc.com, and a theoretical eva luation of peptide toxicity through the Toxinpred platform. It must be highlighted that these servers are based on the eva luation of the isoelectric point, number of residues loaded, and peptide length to determine peptide solubility in water. In addition, the profile of intrinsic solubility was calculated and plotted based on residues where values >1 are highly soluble and <-1 are poorly soluble in solutions at pH 732.
Predicted immunogenicity of the iPPIs
The antigenicity of the peptides was evaluated to predict their safety and immunogenic reactivity. This prediction was done with “Predicting Antigenic Peptides” http://imed.med.ucm.es/Tools/antigenic.pl and VaxiJen servers http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html. Additiona lly, a predictive analysis of T-cell epitopes bound to the major histocompatibility complex I and II was performed using the complete set of alleles available in Immune Epitope Database (IEDB) http://tools.immuneepitope.org/main/.
Results
Sequence analysis of E2-HCV protein and its variability among viral genotypes
The multiple alignment of E2-HCV protein sequences showed highly conserved regions at amino acid positions 501-507, 520-586, and 605-675 among HCV genotypes 1-6; specifi cally, these regions interact with cell receptors such as CD81.
Interaction complexes between the E2 protein of the HCV (E2-HCV) and the CD81 and CD209 cell receptors
From the balanced models carried out through ClusPro, those with the lowest binding energies were selected, which were -1074.4 kcal/mol for the CD81/E2-HCV interaction. (Figure 1) and -1104.2 kcal/mol for the CD209/E2-HCV interaction (Figure 2). The electrostatic potential showed low energies (kcal/(mol*e) in the interaction site of each of the models.
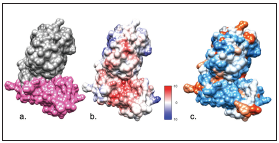
Figure 1 Interaction model generated by ClusPro 2.0® for a. CD81/E2. Observe, in grey, the E2-HCV protein (PDB:4WEB) and, in pink, the CD81 receptor (PDB:5CTX). b. Electrostatic potential, according to Coulomb’s law (the scale varies between -10 and 10 kcal/(mol*e) c. Interactive hydrophobicity surface (blue: hydrophilic amino acids, red-orange: hydrophobic amino acids). Structures visualized in UCSF Chimera®
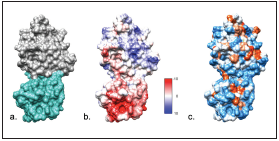
Figure 2 Interaction model generated by ClusPro 2.0® CD209/E2 complex. a. Observe, in grey, domain 2 of the envelop viral protein (PDB:4WEB) and, in light aquamarine green, CD209 cell receptor (PDB: 1K9I). b. Electrostatic potential, according to Coulomb’s law (the scale varies between -10 and 10 kcal/(mol*e) c. Interactive hydrophobicity surface (blue: hydrophilic amino acids, red-orange: hydrophobic amino acids). Structures visualized in UCSF Chimera®.
Peptides derived from the protein-protein interaction (iPPI)
From the CD81/E2-HCV and CD209/E2-HCV interaction mo del, four peptides were obtained from 18 residues with in terface scores of -25.329 and -6.4 REU (Table 3). Given that the server conducts models against both chains of the com plex (receptor and ligand), the iPPI A2 and iPPICD2 peptides were selected because they interact with the viral sequences than with iPPI A1 and iPPI CD1, doing so in relation to the cell receptor, which is why they could interfere on processes specific of the CD81 receptor and CD209 receptor and were discarded as possible candidates.
Table 3 Characteristics of inhibitory peptides of the protein-protein interaction obtained from the Peptiderive in Rosetta online serve
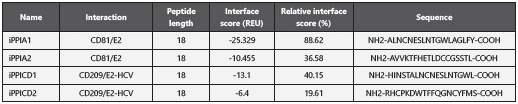
According with the REU scale, molecules with REU below -10 should be considered inhibition candidates. As observed in Table 1, the iPPICD2 obtained a score of -6.4; although it is above -10, it was the candidate with the best score by the Peptiderive server, which is why it was considered in this study
Upon obtaining the peptides, these were again loaded on ClusPro as method to validate the iPPI location. When con ducting the interaction complexes between the iPPIA2 and iPPICD2 peptides and the E2-HCV protein, it was observed that they bind on the same sites where the CD81 and CD209 receptors interacted in the E2-HCV protein. The binding ener gies were -856.6 for the iPPIA2/E2-HCV complex and -961.4 for the iPPICD2/E2-HCV complex (Figure 3). Additionally, it has been demonstrated that it is on residues 412-446 and 519-535 of the E2-HCV protein where the interaction takes place with the cell receptor in vitro studies33,34.
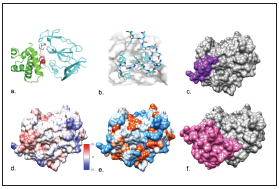
Figure 3 a. binding site of the iPPIA2 (red) to the CD81 complex (green) /E2- HCV (light blue) b. iPPIA2 in light blue and E2-HCV in grey. c. binding site of the iPPIA2C>A(Violet) to the E2-HVC (grey) d. Electrostatic potential according to Coulomb’s law (the scale varies between -10 and 10 kcal/(mol*e) e. Interactive hydrophobicity surface (blue: hydrophilic amino acids, red-orange: hydrophobic amino acids). f. binding site of the CD 81 receptor (pink) to the E2-HVC (Grey). Structures a and b visualized in Rosetta server (https://rosie. rosettacommons.org/). Structures c to f visualized in UCSF Chimera®.
Theoretical physicochemical and cytotoxic properties
According to the results obtained through the theoretical physicochemical and cytotoxic analyses, it was found that the iPPIA2 peptide was poorly soluble in water and can present toxicity, hence, two changes were made in the amino acid chain to improve these parameters. The first modification consisted in designing a peptide at 20 amino acids through Rosetta to try to improve the peptide’s solubility in water. It carried out the addition of a lysine residue in the amino-ter minal region and a threonine residue in the carboxyl-termi nal region. According to this, the iPPIA2,2 peptide improved solubility in water, but continued showing toxic characteris tics. To avoid this toxicity and according to the server’s re commendation, a second modification was made, which con sisted in substituting the cysteine amino acid in position 13 by an alanine. The modified peptide was named iPPIA2C>A. The model of 20 amino acid residues was validated, showing that it continues being a candidate for inhibitory peptide of the CD81/E2-HCV interaction. Again, the physicochemical and toxic properties were calculated, demonstrating that the peptide does not behave as a toxin (Table 2). Replacement of cysteine by alanine in iPPIA2C>A at structural level formed a longer hélix (Figure 4) (Figure 5).
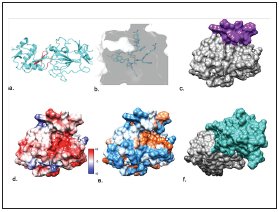
Figure 4.a Binding site of the iPPICD2 (red) to the CD209/E2-HCV complex (light blue). b. iPPICD2 in light blue and E2-HCV in grey. c. binding site of the iPPICD2 (Violet) to the E2-HVC (Grey) d. Electrostatic potential according to Coulomb’s law (the scale varies between -10 and 10 kcal/(mol*e) e. Interactive hydrophobicity surface (blue: hydrophilic amino acids, red-orange: hydrophobic amino acids). f. binding site of the CD 209 receptor (light aquamarine green) to the E2-HVC (Grey). Structures a and b visualized in Rosetta server (https://rosie. rosettacommons.org/). Structures c to f visualized in UCSF Chimera®.
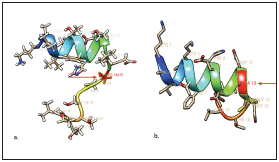
Figure 5 iPPI derived CD81 with and without substitution of the amino acid in position 13. (a.) representation of peptide iPPI A2,20A with the cysteine residue (CYS 184 B.) and (b.) representation of peptide iPPIA2C>A modified with an alanine in position 13. (Structures visualized in UCSF Chimera®.)
According to the results obtained through the theoretical physicochemical and cytotoxic analyses, the iPPICD2 peptide showed low solubility in water; however, it does not behave as a toxin, which is why additional modifications were not made (Table 4).
Table 4 Theoretical physicochemical and cytotoxic properties of the inhibitory peptide of the interaction between CD81/E2.
SN: without modification
M1: Modification 1; a lysine residue in the amino-terminal region and another threonine residue in the carboxyl-terminal region.
M2: Modification 2; substitution of cysteine for alanine in position 13.
*: Given the solubility index, net charge, and isoelectric point of the peptide, solubility in water could be achieved using an organic solvent or a slightly acidic buffer.
Evaluation of the intrinsic solubility profile for both candidate peptides (Figure 6) showed that the solubility score for the iPPIA2C>A peptide was 1.48, demonstrating that this peptide in solutions at pH 7 is soluble as in water. The iPPICD2 pepti de showed a score of 0.23; although it is not highly insoluble due to its proximity to 1, its solubility is reduced in solutions with a pH of 7 and in water.
Predicted antigenicity of the iPPIs
The antigenicity analysis of the peptides using “Predicting Antigenic Peptides” http://imed.med.ucm.es/Tools/antigenic. pl showed that peptides iPPIA1, iPPICD1, and iPPICD2 did not have antigenic determinants in their amino acid sequence, while iPPIA2C>A shows an antigenic determinant of eight re sidues from amino acid 9 to 16 NH2-ETLDACGS-COOH. Vaxi Jen platform showed that iPPIA2C>A and iPPIA1, with scores of 0.29 and 0.38 (<0.5), do not behave as antigens, while iP PICD2 and iPPICD1, with scores of 0.54 and 0.80 (>0.5), likely behave as antigens.
The predicted binding affinity of the peptides to MHC I and II indicated high affinity for MHC II. Regarding MHC I, peptides iPPIA1 and iPPICD1 exhibited low affinity with scores <0.5, while iPPICD2 showed affinity for alleles HLA-A*23:01 HLA-A*24:02 HLA-A*01:01, with scores of 0.63, 0.57, and 0.53, res pectively. Additionally, peptide iPPIA2C>A displayed a pre dicted affinity for allele HLA-B*08:01 (score 0.53).
Discussion
The mechanism of infection through HCV implies interac tion with different cell types. Despite its tropism with liver cells, it has been demonstrated that the virus interacts with lipoproteins and lipids to form lipoviral particles, as well as receptors, like the scavenger receptor type B class I (SR-BI), heparan sulfate, Niemann-Pick C1- Like 1 (NPC1L1), CLDN1 and OCLN, CD36, CD209 and L-SIGN, and tetraspanin CD81 that allow it to recognize and enter its genetic material into the host cell and initiate mechanisms of viral protein replica tion and generation 4,6,8,33,35. This work used CD81 and CD209 receptors as candidates to design the iPPI, given that they have been correlated as high-affinity receptors interac ting directly with the virus to enter the cell and not indirectly, as with other receptors, for example CLDN1 and OCLN8.
It is worth highlighting that interactions with lipoproteins (Apo groups), receptors, and co-cell receptors are mediated principally through the E2-HCV protein, even being functio nal in the absence of the E1 protein-HCV in in vitro tests. However, in living organisms, it has been evidenced that E1- HCV is indispensable as coadjuvant of the E2-HCV protein in recognition and in the subsequent process to carry out mem brane fusion and bring the viral genome inside the cell4,33,34,36.
Given the principal role of the E2-HCV protein in cell recogni tion through the CD81 and CD209 receptors catalogued with high affinity for HCV, interaction complexes were conducted for both CD81/E2-HCV and CD209/E2-HCV proteins, respec tively, to design peptides that inhibit the interaction between these two proteins, suggesting, moreover, that inhibition of this viral protein will permit extrapolating to the rest of the receptors and functions of the E2-HCV protein. According to Miao et al., 20178, binding of the HCV to the CD81 receptor takes place through the long extracellular loop (LEL) that in cludes amino acids 113 to 201, specifically among those resi dues from the CD81 was the protein complex conducted. In turn, Yost et al., (5 mention that the binding site to CD81 in the E2-HCV protein is between residues 412-446 and 502-535, amino acids that interact in the model created in this work. Similarly, these sites would be involved in the interaction with CD209 (Figure 7) (33,37.
Lavie et al. 201438 found that residues Y507, V514, and V515 of the E2-HCV protein are crucial for recognition of the CD81 receptor based on mutagenesis assays. Furthermore, the results of the variability analysis performed in this study showed that residues Y507 and V515 are conserved among all genotypes, in addition to region 520-538, as previously reported (Figure 7c). These results are interesting since the iPPI described in this study could block the interaction on conserved residues among HCV genotypes.
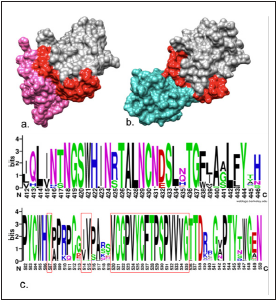
Figure 7 a. CD81/E2 complex. Observe, in pink, amino acids 113-201 of the LEL from the CD81 receptor interacting with residues 412-446 and 519-535 marked in red from the E2-HCV protein (grey). b. CD209/E2-HCV complex. Note, in light aquamarine green, the CD209 receptor in interaction with residues 412-446 and 519-535 marked in red from the E2-HCV protein (grey). Visualized in UCSF Chimera®. c. Multiple alignment of amino acid sequences of the E2-HCV protein from genotypes 1-6. The red box shows residues that are crucial for the interaction with cell receptors, according to literature reports.
The objective of working with peptides is due to increasing evidence on using iPPI as a potential tool for therapeutic use given its safety and tolerability; however, its successful use depends on its target (bearing in mind that computational studies do not completely evaluate the components of which, biologically speaking, the proteins are composed and their immediate environment) and on the modifications made on the peptide39. Some authors mention that modifications on the peptides could improve their stability for use in living organisms, some of these to consider include the forma tion of cyclical peptides through the formation of di-sulfide bridges through cysteines, stabilization of a-helixes, addition of stabilizing chemical groups, among others39,40. According to this, the peptides obtained under the Rosetta web server underwent some modifications, such as seeking to cycle the peptide; however, the peptide’s conditions did not improve (data not shown), hence, modification was only made for the iPPIA2,213C>A peptide to obtain a peptide of 20 amino-acid residues and change in amino acid 13 of a cysteine by an ala nine, Although the size of cysteine is superior to alanine, this does not interfere on the interaction with the E2-HCV protein, but does improve the helix structure of the peptide and avoids its behavior as a toxin. Toxicity prediction using Toxinpred is done based on the peptide composition that is compared aga inst a dataset of toxic and non-toxic peptides retrieved from SwissProt and TrEMBL and not through structural analyses41.
An antigenicity analysis of the four peptides was included to evaluate the safety and reactivity of the peptides with the im mune system of live models. Although peptides iPPICD2 and iPPIA2C>A are derived from cell receptors, iPPIA2C>A showed an antigenic determinant of eight residues predicted by one of the platforms, while VixJen did not predict antigenic behavior for this peptide but only for iPPICD2. These findings show that the same results are not obtained using both platforms and, although these could be considered antigenic, the recognition mechanisms of the organism would prevent an autoimmune response if the peptides were processed intracellularly42. Un like iPPICD2 and iPPIA2C>A, peptides iPPIA1 and iPPICD2 are derived from the E2 protein and would both be expected to display antigenicity; however, only peptide iPPICD1 was pre dicted to behave as an antigen. These results could be useful in the discovery of vaccine candidates or peptides that stimu late an immune response against HCV, or in the development a peptide to standardize diagnostic methods.
Several efforts have been made in the search for peptides against HCV; some authors have sought to inhibit viral pro tein function, and others aimed to inhibit cell receptors or use peptides derived from viral proteins to stimulate the im mune response and serve as neutralizing molecules. Further more, E1 and E2 proteins have been used as targets to inhibit the interaction between the viral protein and the cell since several amino acid residues are highly conserved among ge notypes, suggesting good candidates for this purpose20-23.
Finally, a considerable amount of studies on non peptidic molecules have aimed to inhibit the function or interaction of the E2-HCV protein to diminish the replication percen tage of HCV43-52. Unlike these molecules, the structural and physicochemical characteristics of the peptides make them more effective candidates for inhibition of protein interac tions because they can bind through hot-spot sites from the interaction among proteins, thus, blocking the canoni cal interaction of the target proteins. Another advantage is that they can interact with residues of the protein-protein interfaces, improving the binding specificity and interference in the interaction, which is why the design of the peptides in this work may be considered to conduct in vitro tests53. Computational predictions are important since they provide an initial approximation to the study of biomolecules. These methodologies reduce analytical costs and allow discarding a large number of molecules that could fail in vitro or in vivo assays. All computational approximations are theoretical and each computational tool, including those used here, has its advantages and disadvantages. Furthermore, computational approaches do not consider the biological context in which the peptides and their targets will interact with other organic and inorganic molecules, which could alter the predictions; thus, laboratory assays provide the final results53.
In conclusion, the results obtained in this study showed two peptides iPPIA2C>A and iPPICD2 nontoxic with solubility possible water that could block the interaction cell/virus on conserved residues of the E2 protein among HCV genotypes. It is necessary to highlight that computational approaches do not reflect the results of an in vitro assessment, which is es sential to correlate the computational data. Additionally, the peptides iPPIA1 and iPPICD2, derived from the E2 protein, could be considered vaccine candidates for future studies.