Introduction
The genus Vibrio is naturally found in marine and freshwater habitats and is considered the most diverse marine bacterial genus; this genus is composed of at least 80 species, 12 of which are considered pathogenic to humans1. These species are transmitted to humans through the consumption of contaminated food or water, especially by the consumption of undercooked marine foods, and a smaller number of infections are caused by the contact of open wounds with marine ecosystems1. Vibriosis is defined by different clinical presentations, including self-limiting or extraintestinal gastrointestinal infections, wound infections and cases of sepsis; Nontoxigenic Vibrio cholerae non O1/O139, Vibrio parahaemolyticus, Vibrio vulnificus, Vibrio alginolyticus and Vibrio fluvialis are the species recovered most frequently from these infections1,2.
Currently, vibriosis has gained importance as an infectious disease, since current variations in environmental conditions have increased the population of these species in their ecological niches, thus representing a reserve of potential pathogens for humans2. The report from the Center for Disease Control and Prevention’s and Cholera and Other Vibrio surveillance program (COVIS) showed an increase in annual prevalence of vibriosis from 1996 to 2014 and estimated the occurrence of approximately 1,000-1,500 cases per year3.
Notification of cases of vibriosis is not mandatory in Colombia; however, as a result of the cholera surveillance program, isolates of other Vibrio species have been recovered, which indicates the possible existence of environmental reservoirs of these pathogens and therefore potential outbreak foci, which is a latent threat to the Colombian population4. Hence, the specific, reliable and efficient identification of Vibrio species from the recovered isolates from both clinical and environmental samples leads to better management of the disea- se, as well as an understanding of the distribution of these species in the environment, which in the future will allow propose a strategy for vibriosis surveillance and the projection of possible risk zones for the acquisition of vibriosis5.
The results of commercial phenotypic tests used in the laboratory, such as Vitek, API20E and Microscan, are not reproducible in most cases, generating inconsistent results in diagnoses. The recommendation is to complement the assays with molecular tests using DNA, such as polymerase chain reaction (PCR). Different molecular targets, such as virulence or conserved genes, have been used in PCR for the simultaneous detection of one or more Vibrio species5. Among these, one that is reported for V. alginolyticus is the gyrB gene that encodes subunit B of the gyrase protein6, for V. fluvialis is the membrane binding region of the transcriptional activation domain of toxR, which is divergent in different species of Vibrio7, for V. parahaemolyticus is the tlh gene that encodes thermostable hemolysin that is preserved in all isolates of this species8 and for V. vulnificus is a segment of the vvhA gene that encodes hemolysin9.
In Colombia, there is no active surveillance of vibriosis, however, as a tangential result of the intensive surveillance of cholera10 and a study conducted to search for Vibrio in environmental reservoirs in marine-coastal systems during 2018-2019, Vibrio isolates have been recovered, including nontoxigenic V. cholerae non O1/O139, V. parahaemolyticus, V. vulnificus, V. alginolyticus and V. fluvialis, which have been identified by CHROMagar™ Vibrio and with automated and semiautomated systems such as Vitek and API20E11. However, the incongruous results between these three methods have led us to develop a multiplex PCR for the identification of V. alginolyticus, V. fluvialis, V. parahaemolyti- cus and V. vulnificus from pure colonies previously characterized by the methods mentioned above to confirm the species of the microorganisms recovered from clinical samples and samples from bodies of water.
Materials and Methods
Bacterial isolates
A total of 192 isolates were included in this study, which included two reference strains of toxigenic V. cholerae O1, non- toxigenic V. cholerae non O1/O139, V. parahaemolyticus, V. vulnificus, V. alginolyticus and V. fluvialis, which were re- covered from water samples with three different salinity conditions (marine, estuarine and continental) from Instituto de Investigaciones Marinas y Costeras José Benito Vives de Andréis (INVEMAR) and from different clinical samples (stool samples, blood cultures, wounds and cerebrospinal fluids) from Instituto Nacional de Salud (INS). This study additionally included isolates of other genera.
DNA extraction
All isolates were grown on Brain Heart Infusion (BHI) agar overnight at 37 ºC. Deoxyribose Nucleic Acid (DNA) extraction was performed by boiling a suspension of 2-3 bacterial colonies in 100 µl Tris HCl 0.1 N at 100oC for 15 minutes to lyse the bacteria. The suspension was then centrifuged for 2 minutes at 12000 rpm to sediment the cell debris, and the supernatant was collected in another tube for use as the DNA template and was stored at 20 ºC until use. DNA was extracted from the reference strains with the Qiagen mini DNA kit (QIAamp DNA Mini Kit) following the protocol and recommendations of the manufacturer to determine specificity.
Estandardization Multiplex PCR
Genetic targets reported in previous studies were selected for the identification of V. alginolyticus, V. fluvialis, V. parahaemolyticus and V. vulnificus (Table 1a). The reported amplification sizes between each species were sought to have minimum differences of 100 bp for proper separation in agarose gels. These parameters were met only with the gyrB, tlh and toxR reported genes. Therefore, we designed primers in a region of the gene that encodes hemolysin vvhA that amplified a fragment of 710 bp.
Table 1. Primers selected to evaluate the specificity of the multiplex PCR for the identification of V. alginolyticus, V. parahaemolyticus, V. fluvialis and V. vulnificus.
Table 1a Primers for the identification of V. alginolyticus, V. parahaemolyticus, V. fluvialis and V. vulnificus.
| Vibrio spp. | Target gene | Sequence 5-3´ | Size& | Mt (°C) | References |
|---|---|---|---|---|---|
| V. alginolyticus | gyrB- F | GAGAACCCGACAGAAGCGAAG | 337 bp | 61.8 | 6 |
| gyrB- R | CCTAGTGCGGTGATCAGTGTTG | 62.1 | |||
| V. fluvialis | toxR-F | GACCAGGGCTTTGAGGTGGACGAC | 217 bp | 67.8 | 7 |
| toxR-R | AGGATACGGCACTTGAGTAAGACTC | 63.0 | |||
| V. parahaemolyticus | tlh -F | AAAGCGGATTATGCAGAAGCACTG | 450 bp | 61.0 | 8 |
| tlh-R | GCTACTTTCTAGCATTTTCTCTGC | 59.3 | |||
| V. vulnificus | vvhA- F | AATCGGCAACGTCAGATGGT | 710 bp | 57.3 | This study |
| vvhA-R | GCCGTAAACCGAAAACAGCG | 59.3 |
& Expected size band
Initially, the amplification conditions were optimized for each gene separately and then for all genes together for multiplex PCR. The specificity of each of the primers was evaluated with isolates of the four species confirmed by three phenotypic methods, CHROMagar™ Vibrio, Vitek and API20E, as well as DNA of taxonomically related genera that can be found cohabiting with Vibrio spp. in ecological niches or coinfecting clinical samples (Table 1b). The resulting fragments were sequenced and confirmed by Basic Local Alignment Search Tool (BLAST). Isolates identified as nontoxigenic V. cholerae non O1/O139 by phenotypic methods too were confirmed by PCR following previously published protocols12.
Table 1b: Isolates selected to evaluate the specificity of the multiplex PCR
| Bacterial species | Source |
|---|---|
| Vibrio parahaemolyticus | INS strain bank |
| Vibrio alginolyticus | INVEMAR |
| Vibrio vulnificus | INS strain bank |
| Vibrio fluvialis | INS strain bank |
| Vibrio cholerae No 01/0139 | INS strain bank |
| Vibrio cholerae 01/0139 | INS strain bank |
| Vibrio mimicus | INVEMAR |
| Aeromona salmonicida | INVEMAR |
| Shewanella algae | INVEMAR |
| Photobacterium damselae | INVEMAR |
| Sphingomonas paucimobilis | INVEMAR |
| Escherichia coli | INS strain bank |
| Salmonella Enteritidis | INS strain bank |
| Listeria monocytogenes | INS strain bank |
| Yersinia enterocolitica | INS strain bank |
The PCR master mix contained 1X PCR buffer, 3 µM MgCl , 2U Taq polymerase (Invitrogen), 0.8 µM dNTP mixture (Promega), 0.5 µM each primer (Invitrogen), 3.6 µL molecular grade water and 5 µL DNA for a final volume of 25 µL. The thermocycler conditions were programmed as follows: initial denaturation at 95 ºC for 3 minutes, followed by 30 denaturation cycles at 95 ºC for 30 seconds, annealing at 58 ºC for 30 seconds, elongation at 72 ºC for 30 seconds and 1 final elongation cycle at 72 ºC for 5 minutes; the products were run on a 2% agarose gel for 35 minutes at 110 V and 400 mA. The expected fragment sizes were 217 bp for V. fluvialis, 337 bp for V. alginolyticus, 450 bp for V. parahaemolyticus and 710 bp for V. vulnificus.
The detection limit for each target gene was determined separately with different DNA dilutions of the control strains, starting from a concentration of 35 ng/µL up to 7 1:2 serial dilutions, with concentrations of 17.5 ng/µl, 8.75 ng/µl, 4.37 ng/µl, 2.18 ng/µl, 1.09 ng/µl, 0.54 ng/µl and 0.27 ng/µl. The DNA concentration was evaluated with a Thermo Scienti- fic equipment™ NanoDrop 2000.
Results
The primers selected were first evaluated in a monoplex PCR with the reference strains and evaluated with isolates of the other four species. The specificity for each species was confirmed, observing the amplification of the expected band of 710 bp for V. vulnificus, 450 bp for V. parahaemolyticus, 337 bp for V. alginolyticus and 217 bp for V. fluvialis and without amplification of DNA from bacteria representative of another genus (n=7) (Figure 1a). The amplified sequences of each of the products were 100% specific to the respective genes deposited in GenBank (data not shown). The separate detection limits for the tlh, vvhA, and gyrB genes was 1.09 ng/µl DNA, while that for toxR was 2.18 ng/µl DNA (Figure 1b). Once the specificity was confirmed, the conditions for the multiplex Vibrio PCR were established.
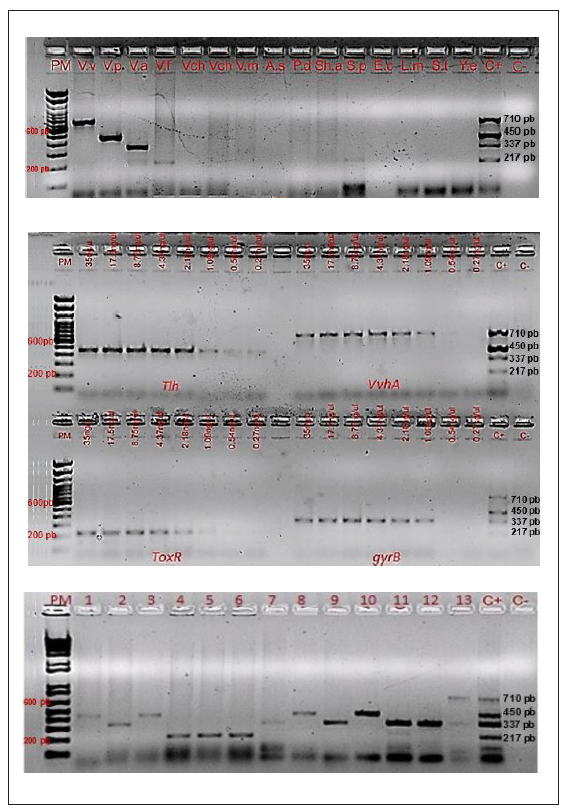
Figure 1 Specificity, detection limit and multiplex PCR for Vibrio spp. Figure 1a. Specificity of multiplex PCR for Vibrio spp. Lanes 2-16: V.v: V.vulnificus (710bp), V.p: V. parahaemolyticus (450bp), V.a: V. alginolyticus (337bp), V.f: V.fluvialis (217bp), Vch: V. cholerae, V.m: V.mimicus, A.s: Aeromonas salmonicida, P.d: Photobacterium damselae, Sh.a: Shewanella algae, S.p: Sphingomonas paucimobilis, E.c: Escherichia coli, L.m: Listeria monocytogenes, S.E: Salmonella Enteritidis, Y.e: Yersinia enterocolitica. Figure 1b. Detection limit of each primer at different DNA concentrations in multiplex PCR for Vibrio spp. Lanes 2-17: concentrations of DNA. Genes tlh (V.p: V. paraemolyitcus-450 bp), vvhA (V. v: V. vulnificus-710 bp), toxR (V.f: V. fluvialis-217 bp). gyrB (V.a: V. alginolyticus-337 bp). Figure 1c. Multiplex PCR for Vibrio spp. Lanes 1-13: clinical and environmental isolates tested by PCR-Multiplex. PM: Molecular weight. C+ is the control sample used to verify the amplified fragments: V. vulnificus (710bp), V. parahaemolyticus (450bp), V. alginolyticus (337bp), V. fluvialis (217bp).
Of the 192 isolates analyzed, 124 (64.6%) were confirmed as one of the four species by multiplex Vibrio PCR: V. para- haemolyticus n= 54, V. alginolyticus n= 38, V. vulnificus n=7 and V. fluvialis n= 25. The remaining 68 isolates (35.4%) were negative by multiplex PCR and were classified as other Vibrio spp. or non-Vibrio according to the phenotypic results obtained previously (Table 2, Figure 1c).
Table 2 Distribution of 192 isolates identified by multiplex PCR
| Species | Environmental (%) | Clinical (%) | Global Distribution n (%) |
|---|---|---|---|
| V. parahaemolyticus | 43 (79.6) | 11(20.4) | 54(28.1) |
| V. alginolyticus | 36 (94.7) | 2 (5.3) | 38(19.8) |
| V. fluvialis | 16 (64.0) | 9(36.0) | 25(13.2) |
| V. vulnificus | 6(85.7) | 1(4.3) | 7(3.64) |
| Subtotal | 101 | 23 | 124 |
| No amplification | 61 | 7 | 68 |
| Total | 162 | 30 | 192 |
In total, the phenotypic results and the multiplex Vibrio PCR had correlating results in 139 of 192 isolates (72.4%); the species with the highest correlation with respect to the phenotypic identification was V. parahaemolyticus, and the lowest correlation was observed with V. vulnificus.
Discussion
The standardization of a multiplex PCR for the simultaneous identification of four Vibrio species was successful, taking into account specificity against genetic targets and the use of false positives with other related genera, both from the ecological niches in which Vibrio naturally occur and in clinical cases where Vibrio could be misidentified due to the physiopathology of the clinical case; this was further confirmed by sequencing the amplified products. The specificity evident in our PCR was expected compared to the results of pre- vious studies using the same primers 13,14, additionally, we report that the primers for V. vulnificus designed in this study showed great detection capacity and specificity.
While other reported studies have suggested the identification of multiple Vibrio species by PCR, they have focused on species of clinical importance in a particular geographical area, as described by Weit et al., where instead of V. fluvialis, V. mimicus is identified14 or by Yin et al., who identified V. alginolyticus, V. parahaemolyticus and V. vulnificus in clinical and environmental samples15. In Colombia, the targeted species of this study are those recovered mainly from clinical samples, which prompted us to standardize and implement a useful multiplex PCR applicable to the real epidemiology of our country, achieving faster identification than with monoplex PCR.
The predominance of V. parahaemolyticus and V. alginolyticus in environmental samples submitted by INVEMAR is expected considering that these have already been reported as predominant species in aquatic environments15 and are coincidentally prevalent in the COVIS annual summaries as causative agents of clinical cases if vibriosis in the United States, which shows the pathogenic potential of these species. In this way, the Caribbean Colombian coast is an environmental reservoir for V. alginolyticus, while the Pacific coast is a reservoir for V. parahaemolyticus, and are potential sources of transmission through mariculture activities or consumption of contaminated water, which is supported by studies indica- ting that V. parahaemolyticus infections have been strongly associated with shellfish ingestion and occur more often on the Pacific coast of South America16 than on the Caribbean coast. The isolation of Vibrio spp. was performed in oysters, and while all 3 species were observed, there was a higher prevalence of V. alginolyticus in oysters17.
Thus, the results of this study show that V. cholerae is not the only pathogenic species of Vibrio of clinical interest in Colombia, as it was observed that there are environmental reservoirs in the country for other species that cause di- sease. Several reports mention that these species of Vibrio have emerged in different locations worldwide over time, including in Latin America and have been associated with gastrointestinal and extraintestinal infections and even with outbreaks of foodborne illnesses18.
It is also important to mention the observed association between the phenotypic identification previously made by the research group with respect to the genotypic analysis performed in this study, in which 70% agreement is evident. Currently, the advantages of molecular methods over phenotypic tests are already evident as phenotypic tests take longer to generate a result and are very variable, as they depend on aspects such as the origin of the samples (clinical or environmental), as studies on V. parahaemolyticus suggest19,20. Conversely, detection using molecular methods, such as PCR targeting specific genes, provides a sensitivity and specificity that is approximately 100% with respect to phenotypic test that range in sensitivity and specificity from 70-90%; although phenotypic identification may provide presumptive characterization, confirmation by molecular methods is suggested for a more accurate species identification19,20.
Multiplex PCR was developed for the detection of V. alginolyticus, V. fluvialis, V. parahaemolyticus and V. vulnificus, and was determined to be reliable, specific, sensitive and have a high discriminatory capacity for identifying isolates reco- vered from both clinical and environmental samples, with 70% agreement with phenotypic methods.
The target species of Vibrio were identified, as well as their geographical distributions in different ecological niches consisting of coastal areas of the country. These species account for approximately 60% of the total microorganisms present in INVEMAR environmental samples and 72% of the total surveillance samples.
Considering all the results of this study, we can suggest the implementation of continuous monitoring and surveillance of these bacterial species in environmental reservoirs, as they could be considered agents of potential outbreaks in Colombia, and thus we anticipate the possible increase in the population of Vibrio spp. in order to propose strategies for the vigilance and containment of the diseases these organisms cause.
Ethical considerations
Protection of human and animal subjects. The authors declare that no experiments were performed on humans or animals for this investigation. The research was approved by the ethics committee of each of the participating centers and the Universidad Libre -Cali Campus
Right to privacy and informed consent. The authors declare that no data that enables identification of the patients appears in this article.















