Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista de Salud Pública
versão impressa ISSN 0124-0064
Rev. salud pública v.12 n.3 Bogotá maio/jun. 2010
Juan C. Rozo-Anaya1 y Wellman Ribón2
1 Grupo de Micobacterias, Instituto Nacional de Salud. Bogotá. Centro Colombiano de Investigación en Tuberculosis-CCITB. Universidad de Pamplona. Colombia.juancarlosrozoanaya@hotmail.com
2 Grupo de Micobacterias, Instituto Nacional de Salud. Bogotá. Centro Colombiano de Investigación en Tuberculosis-CCITB. Escuela de Bacteriología, Universidad Industrial de Santander, Bucaramanga. Colombia. wellmanribon@yahoo.es
Received 27th July 2009/Sent for Modification 1th April 2010/Accepted 27th June 2010
ABSTRACT
Objective The present work studied molecular typing methods used for Mycobacterium tuberculosis characterization in order to learn about their advantages, disadvantages and discrimination power as regards the implementation of tuberculosis surveillance and control programs.
Methods To analyze the discrimination power of each method we studied articles that included Hunter-Gaston discrimination index (HGDI) values or data allowing their calculation.
Results The highest discrimination power was registered for LM-PCR followed by FLiP and 15-loci MIRU. The most frequently used methods showed an HGDI of 0.9491, 0.9519 and 0.8630 for 12-loci MIRU, RFLP-IS6110 and spoligotyping, respectively.
Conclusion M. tuberculosis isolates molecular characterization requires at least two molecular markers to discriminate non related isolates, as well as previous analysis to their implementation.
Key Words: Tuberculosis, Mycobacterium tuberculosis, molecular epidemiology, genotype, DNA, Polymorphism, Restriction Fragment Length (source: MeSH, NLM).
RESUMEN
Objetivo En el presente trabajo se estudiaron las metodologías de tipificación molecular empleadas para caracterizar Mycobacterium tuberculosis con el objetivo de conocer las ventajas, desventajas y poder discriminatorio para ser consideradas al momento de la implementación en los programas de vigilancia y control de la tuberculosis.
Métodos Para el análisis del poder discriminatorio de cada metodología se estudiaron los artículos que suministraban el valor del Hunter-Gaston discrimination index (HGDI) ó los datos que permitían su determinación.
Resultados Se documentó que el LM-PCR tiene una mayor capacidad discriminatoria seguida de FLiP y MIRU de 15 loci. Las metodologías más comúnmente empleadas mostraron un HGDI de 0.9491, 0.9519 y 0.8630 para MIRU de 12 loci, RFLP-IS6110 y spoligotyping respectivamente.
Conclusión La caracterización molecular de aislamientos de M. tuberculosis requiere mínimo el análisis de al menos dos marcadores moleculares para discriminar aislamientos no relacionados y la necesidad de realizar análisis previos a la implementación de estas metodologías.
Palabras Clave: Tuberculosis, Mycobacterium tuberculosis, epidemiologia molecular, genotipo, ADN, polimorfismo de longitud del fragmento de restricción (fuente: DeCS, BIREME).
Tuberculosis (TB) is an infecto-contagious disease caused by Mycobacterium tuberculosis complex that affects around a third of the world's population (1). M. tuberculosis genome is markedly homogeneous. The species of M. turberculosis, M. africanum, M. bovis, M. bovis BCG, M. caprae, M. pinnipedii, M. microti and M. canettii are genetically related. The presence of mutations in drug interaction places in genes rpoB (rifampicin), inhA, KatG, ahpC (izoniacid), rrs, rpsl (streptomycin), embB (ethambutol) and gyrA y gyrB (quinolones) results in resistance to these drugs producing a phenotypical change (2). This fact gave way to the development of methods capable of differentiating among this species isolates. Various techniques were used for the differential identification of M. tuberculosis, among them, those for determining unusual drug resistance, serotyping, multilocus enzyme electrophoresis (3), biochemical heterogeneity and phagus typing (4), being the latest one the standard method used until the late 1980's that presented however considerable disadvantages such as the low amount of identified phagotypes and its laboriousness.
The development of new molecular methods for M. tuberculosis genetic characterization has greatly contributed to the understanding of the transmission dynamics and pathogenesis of the disease (5,6). Today, the typing standard method for M. tuberculosis complex species is the restriction fragment length polymorphism (RFLP) -IS6110.
Molecular typing methods for M. tuberculosis complex are grouped in genomic methods for DNA studies such as RFLP IS6110 (7-10), polymorphic GCrich sequence (PGRS) (11-13) analysis and pulsed-field gel electrophoresis (PFGE) on the one hand, and on the other hand, DNA-specific sequence amplification methods using polymerase chain reaction such as spoligotyping (spacer oligonucleotide typing) (14-18), ligation-mediated PCR (LM-PCR) (19,20), double repetitive element PCR (DRE-PCR) (21), fast ligation mediated PCR (FliP) (22), fluorescent amplified-fragment length polymorphism (FAFLP) (23,24), my-cobacterial interspersed repetitive unit (MIRU) (25) analysis, amplification and sequencing of single nucleotide polymorphism (SNP), amplification of exact tandem repeats (ETR) and Queen's University Belfast (QUB) polymorphism of variable number tandem repeat (VNTR) (Table 1).
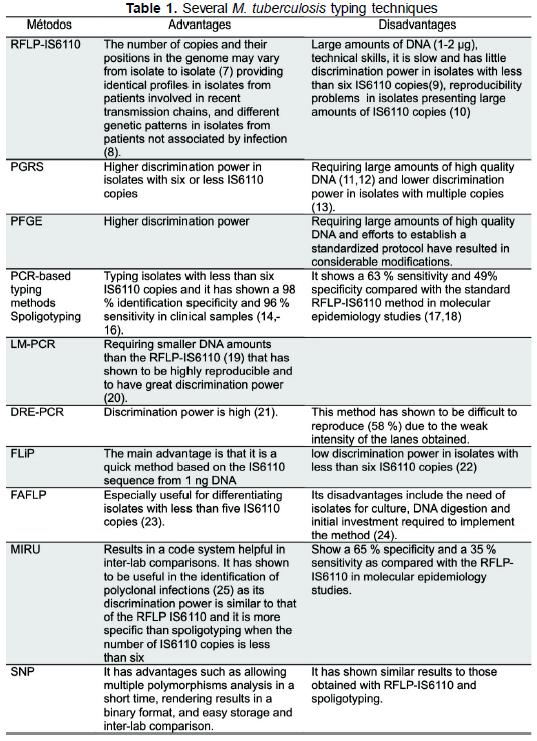
The characteristics, advantages and disadvantages of the different methods used for M. tuberculosis complex molecular typing always pose a problem at the time of choosing the most adequate. We undertook a review of literature by determining and analyzing the discrimination power for each method or combination of methods through the Hunter-Gaston discrimination index (HGDI) (26) as selection criterium at the time of implementing molecular methods in transmission studies because this is the first factor to consider at the initial stage of choosing a method (27).
Publications were searched in PubMed, Blackwell-synergy, ScienceDirect and EBSCOhost electronic data bases from March 2007 to August 2008 including the terms "Mycobacterium tuberculosis complex", "RFLP-IS6110", "spoligotyping", "MIRU", "VNTR", "Pulsed Field Gel Electrophoresis" "Direct repetition", "DRE-PCR", "Fluorescent amplifield fragment leng polymorphism", "FLip", "Genotyping AND Mycobacterium", "SNP", "LM-PCR", "HGDI AND Mycobacterium".
The selection criterium aimed at publications with titles related to M. tuberculosis complex genotyping and choosing those related to epidemiology and molecular characterization that included the Hunter-Gaston discrimination index value (HGDI) or data on the number of groupings and the number of isolates grouped in each of them to enable HGDI determination.
Our search exhibit as LM-PCR showed the highest HGID (0.9980), followed by FLiP (HGID 0.9945) and 15-loci MIRU (0.9620). RFLP-IS6110, the standard method used in M. tuberculosis molecular epidemiology, showed an average HGID of 0.9519. The combination of two or more methods showed higher HGDI values than those found when analyzing each method separately; these values were equal or close to one (28-54) (Table 2).
The different typing methods show discrimination powers expressed in a wide range of HGDI values that go from 0,6044 to 0,9985. A base HIGD value of >0,95 is required to differentiate among related organisms (26), and previous studies of each method should be undertaken in the development of M. tuberculosis complex molecular epidemiology research.
The analysis of HGDI average values for each method under study showed that spoligotyping has an HGDI average of 0.8630; we found values ranging from 0.6582 to 0.9817, the lowest of them resulting from the analysis of isolates obtained in Nigeria and the highest in the Netherlands, which shows the diversity of circulating strains in these places probably due to population migrations in each area.
Kremer et al (55) studied a group of 90 M. tuberculosis complex strains and determined the discrimination power based on the amount of different patterns obtained by typing them using the methods under study (RFLP-IS6110, mixedlinker PCR (LM-PCR), and arbitrarily primed PCR (APPCR), RFLP-PGRS, DRE-PCR, spoligotyping, VNTR, RFLP y RFLP-IS1081). They reported that for epidemiological research the methods of choice should be RFLP-IS6110 and LM-PCR, as they show a higher number of distinct patterns (84 and 81 patterns, respectively) (23).
Clinical isolates characterization at the Instituto Nacional de Salud de Colombia resulted in 11 % of them with less than four IS6110 copies (47), contrasting with the finding reported by Asgharzadeh 2006, who found 8.6 % of isolates with less than six copies (43). Due to the low number of IS6110 copies, a clear disadvantage of this method, complementary methods have appeared such as MIRU, that combined with spoligotyping, provide a higher discrimination power than that one obtained by using a single method (56). MIRU is a recently developed molecular typing method that has the greatest acceptance at the moment, as it shows an adequate balance between variability, an essential feature to differentiate among non related isolates, and molecular marker stability (57) with intermediate sensitivity and specificity as compared with spoligotyping sensitivity and RFLP-IS6110 specificity (58). Like spoligotyping (59), this method has shown to be useful for typing M. bovis species that generally present few copies of IS6110 sequence, and it has a greater discrimination power as compared with spoligotyping (60). However, its standardization has taken several years in a process where
main changes refer to the primers used to enhance amplified product specificity, the magnesium chloride concentration in PCR mixture, the annealing temperature and even the loci analyzed (61), including a 24 loci, that have shown the same or greater discrimination power than RFLP-IS6110. This method has been the only one enabling discrimination of isolates with no IS6110 insertion sequences, excelling, therefore, spoligotyping and PGRS (62).
It is important to note that some methods are seldom used in genotyping and there are few publications reporting results from their implementation: FAFLP and LM-PCR data are reported in only one publication complying with our inclusion criteria. Analysis of HGDI average values in methods included in more than one publication shows that 15-loci MIRU presents the highest discrimination power (0.9620), followed by RFLP-IS6110 (0.9519), 12-loci MIRU (0.9491) and spoligotyping (0.8630) (Table 2). The average HGDI for RFLP-IS6110, excluding the 0.6044 HGDI reported by Dahle 2005, is 0.9808, this being the highest discrimination power agreeing with results reported by Kremer 2005 who found the highest discrimination power in RFLP-IS6110 (HGDI: 0.997) followed by MIRU (HGDI 0.995). Spoligotyping was not included in the analysis done by Kremer 2005 (20).
Our review methodology enabled us to establish the low number of publications determining discrimination power of molecular methods used in M. tuberculosis complex characterization (28 publications from 1,352). This was a limitation in the development of the present study as it has been in other review (44).
Based on the data analyzed in our study we can conclude that M. tuberculosis complex isolates molecular characterization requires at least two molecular markers to discriminate non related isolates as reported by Barlow 2001, who with combination of RFLP-IS6110 and VNTR- ETR obtained a higher HGID (0.988) than the one reported for each method taken separately. Spoligotyping reaches a higher discrimination power (HGDI 0.97) in the characterization of M. tuberculosis complex members by increasing spacing sequences from 43 to 65 (63), as happens with RFLP-IS6110 that provides greater evidence of epidemiological relation between two isolates when combined with other methods.
Implementation of molecular methods for M. tuberculosis complex typing requires an analysis of their advantages and disadvantages as regards tuberculosis surveillance and control programs because their use allows for an extra 52 % detection of epidemiological relation among patients (64) and contributes to better understanding TB transmission dynamics.
The results obtained by our review evidence that laboratories should determine the discrimination power for each molecular method to enable a better selection, implementation and combination according to the specific conditions of each laboratory and the particular features of the geographic region and to guarantee their reproducibility and discrimination power.
Acknowledgements: This study was supported by the Instituto Nacional de Salud (INS) and the Centro Colombiano de Investigación en Tuberculosis (CCITB).
1. Organización mundial de la salud OMS. Plan mundial para detener la tuberculosis 2006-2015. Actuar para salvar vidas. Hacia un mundo sin tuberculosis. Ginebra: Organización Mundial de la Salud; 2006. p. 35. [ Links ]
2. Musser JM. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev 1995;8:496-514. [ Links ]
3. Feizabadi MM, Robertson ID, Cousins DV, Hampson DJ. Genomic analysis of Mycobacterium bovis and other members of the Mycobacterium tuberculosis complex by isoenzyme analysis and pulsed-field gel electrophoresis. J Clin Microbiol 1996;34:1136-42. [ Links ]
4. Kalantri S, Pai M, Pascopella L, Riley L, Reingold A. Bacteriophage- based tests for the detection of Mycobacterium tuberculosis in clinical specimens: a systematic review and metaanalysis. BMC Infect Dis 2005;5:59. [ Links ]
5. Kato-Maeda M, Small PM. How molecular epidemiology has changed what we know about tuberculosis. West J Med 2000;172:256-9. [ Links ]
6. van Soolingen D, De Haas, Kremer K. Restriction fragment length polymorphism (RFLP) typing of mycobacteria. National Institute of Public Health and Environment. Bilthoven. The Netherlands; 2002. [ Links ]
7. van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 1993;31:406-9. [ Links ]
8. Aranaz A, Liebana E, Gomez-Mampaso E, Galan JC, Cousins D, Ortega A, et al. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int J Syst Bacteriol 1999;493:1263-73. [ Links ]
9. Gutacker MM, Smoot JC, Migliaccio CA, Ricklefs SM, Hua S, Cousins DV, et al. Genome-wide analysis of synonymous single nucleotide polymorphisms in Mycobacterium tuberculosis complex organisms: resolution of genetic relationships among closely related microbial strains. Genetics 2002;162:1533-43. [ Links ]
10. Nakamura Y, Obase Y, Suyama N, Miyazaki Y, Ohno H, Oka M, et al. A small outbreak of pulmonary tuberculosis in non-close contact patrons of a bar. Intern Med 2004;43:263-7. [ Links ]
11. Asgharzadeh M, Kafil HS. Current trends in molecular epidemiology studies of Mycobacterium tuberculosis. Biotechnol. Mol Biol Rev 2007;2:108-15. [ Links ]
12. Doroudchi M, Kremer K, Basiri EA, Kadivar MR, Van SD, Ghaderi AA. IS6110-RFLP and spoligotyping of Mycobacterium tuberculosis isolates in Iran. Scand J Infect Dis 2000;32:663-8. [ Links ]
13. Bauer J, Andersen AB, Kremer K, Miorner H. Usefulness of spoligotyping To discriminate IS6110 low-copy-number Mycobacterium tuberculosis complex strains cultured in Denmark. J Clin Microbiol 1999;37:2602-6. [ Links ]
14. Kodmon C, Niemann S, Lukacs J, Sor E, David S, Somoskovi A. Molecular epidemiology of drugresistant tuberculosis in Hungary. J Clin Microbiol 2006;44:4258-61. [ Links ]
15. Gori A, Bandera A, Marchetti G, Degli EA, Catozzi L, Nardi GP, et al. Spoligotyping and Mycobacterium tuberculosis. Emerg Infect Dis 2005;11:1242-8. [ Links ]
16. Baker LV, Brown TJ, Maxwell O, Gibson AL, Fang Z, Yates MD, et al. Molecular analysis of isoniazid-resistant Mycobacterium tuberculosis isolates from England and Wales reveals the phylogenetic significance of the ahpC -46A polymorphism. Antimicrob.Agents Chemother 2005;49:1455-64. [ Links ]
17. Scott AN, Menzies D, Tannenbaum TN, Thibert L, Kozak R, Joseph L, et al. Sensitivities and specificities of spoligotyping and mycobacterial interspersed repetitive unit-variablenumber tandem repeat typing methods for studying molecular epidemiology of tuberculosis. J Clin Microbiol 2005;43:89-94. [ Links ]
18. Supply P, Magdalena J, Himpens S, Locht C. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol Microbiol 1997;26:991-1003. [ Links ]
19. Prod'hom G, Guilhot C, Gutierrez MC, Varnerot A, Gicquel B, Vincent V. Rapid discrimination of Mycobacterium tuberculosis complex strains by ligation-mediated PCR fingerprint analysis. J Clin Microbiol 1997;35:3331-4. [ Links ]
20. Kremer K, Arnold C, Cataldi A, Gutierrez MC, Haas WH, Panaiotov S, et al. Discriminatory power and reproducibility of novel DNA typing methods for Mycobacterium tuberculosis complex strains. J Clin Microbiol 2005;43:5628-38. [ Links ]
21. Mostrom P, Gordon M, Sola C, Ridell M, Rastogi N. Methods used in the molecular epidemiology of tuberculosis. Clin Microbiol Infect 2002;8:694-704. [ Links ]
22. Reisig F, Kremer K, Amthor B, van SD, Haas WH. Fast ligation-mediated PCR, a fast and reliable method for IS6110-based typing of Mycobacterium tuberculosis complex. J Clin Microbiol 2005;43:5622-7. [ Links ]
23. Sims EJ, Goyal M, Arnold C. Experimental versus in silico fluorescent amplified fragment length polymorphism analysis of Mycobacterium tuberculosis: improved typing with an extended fragment range. J Clin Microbiol 2002;40:4072-6. [ Links ]
24. Mortimer P, Arnold C. FAFLP: last word in microbial genotyping? J Med Microbiol 2001;50:393-5. [ Links ]
25. Garcia DV, Alonso RN, Andres S, Ruiz MJ, Bouza E. Characterization of clonal complexity in tuberculosis by mycobacterial interspersed repetitive unit-variable-number tandem repeat typing. J Clin Microbiol 2005;43:5660-4. [ Links ]
26. Hunter P,Gaston M. Numerical Index of the Discriminatory Ability of Typing Systems: an Application of Simpson's Index of Diversity. J Clin Microbiol 1988;26:2465-6. [ Links ]
27. Gaston MA, Hunter PR. Efficient selection of tests for bacteriological typing schemes. J Clin Pathol 1989;42:763-6. [ Links ]
28. Skuce RA, McCorry TP, McCarroll JF, Roring SM, Scott AN, Brittain D. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 2002;148: 519-28. [ Links ]
29. Kassama Y, Shemko M, Shetty N, Fang Z, Macintire G, Gant V, et al. An improved fluorescent amplified fragment length polymorphism method for typing Mycobacterium tuberculosis. J Clin Microbiol 2006;44:288-9. [ Links ]
30. Burger M, Raskin S, Brockelt SR, Amthor B, Geiss HK, Haas WH. DNA fingerprinting of Mycobacterium tuberculosis complex culture isolates collected in Brazil and spotted onto filter paper. J Clin Microbiol 1998;36:573-6. [ Links ]
31. Banu S, Gordon SV, Palmer S, Islam MR, Ahmed S, Alam KM. Genotypic analysis of Mycobacterium tuberculosis in Bangladesh and prevalence of the Beijing strain. J Clin Microbiol 2004;42:674-82. [ Links ]
32. Evans JT, Hawkey PM, Smith EG, Boese KA, Warren RE, Hong G. Automated high-throughput mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis strains by a combination of PCR and non denaturing high-performance liquid chromatography. J Clin Microbiol 2004;42:4175-80. [ Links ]
33. Godreuil S, Torrea G, Terru D, Chevenet F, Diagbouga S, Supply P, et al. First molecular epidemiology study of Mycobacterium tuberculosis in Burkina Faso. J Clin Microbiol 2007;45:921-7. [ Links ]
34. Chin PJ, Jou R. A modified automated high-throughput mycobacterial interspersed repetitive unit method for genotyping Mycobacterium tuberculosis. Diagn Microbiol Infect Dis 2005;53: 325-7. [ Links ]
35. Maes M, Kremer K, van Soolingen D, Takiff H, de Waard J. 24-Locus MIRU-VNTR genotyping is a useful tool to study the molecular epidemiology of tuberculosis among Warao Amerindians in Venezuela. Tuberculosis 2008;88:490-4. [ Links ]
36. Sun YJ, Bellamy R, Lee AS, Ng ST, Ravindran S, Wong SY, et al. Use of mycobacterial interspersed repetitive unit-variable-number tandem repeat typing to examine genetic diversity of Mycobacterium tuberculosis in Singapore. J Clin Microbiol 2004;42:1986-93. [ Links ]
37. Haas WH, Bretzel G, Amthor B, Schilke K, Krommes G, Rusch-Gerdes S, et al. Comparison of DNA fingerprint patterns of isolates of Mycobacterium africanum from east and west Africa. J Clin Microbiol 1997;35:663-6. [ Links ]
38. Dahle UR, Nordtvedt S, Winje BA, Mannsaaker T, Heldal E, Sandven P, et al. Tuberculosis in contacts need not indicate disease transmission. Thorax 2005;60:136-7. [ Links ]
39. Durmaz R, Gunal S, Yang Z, Ozerol IH, Cave MD. Molecular epidemiology of tuberculosis in Turkey. Clin Microbiol Infect 2003;9:873-7. [ Links ]
40. Garcia MR, Rodriguez JC, Navarro JF, Samper S, Martin C, Royo G. Molecular epidemiology of tuberculosis in Elche, Spain: a 7-year study. J Med Microbiol 2002;51:273-7. [ Links ]
41. Martinez M, Garcia D, Alonso VM, Andres S, Bouza E, Cabezas T, et al. Impact of laboratory cross-contamination on molecular epidemiology studies of tuberculosis. J Clin Microbiol 2006;44:2967-9. [ Links ]
42. van der Zanden AG, Kremer K, Schouls LM, Caimi K, Cataldi A, Hulleman A, et al. Improvement of differentiation and interpretability of spoligotyping for Mycobacterium tuberculosis complex isolates by introduction of new spacer oligonucleotides. J Clin Microbiol 2002;40:4628-39. [ Links ]
43. Asgharzadeh M, Shahbabian K, Majidi J, Aghazadeh AM, Amini C, Jahantabi AR, et al. IS6110 restriction fragment length polymorphism typing of Mycobacterium tuberculosis isolates from East Azerbaijan Province of Iran. Mem Inst Oswaldo Cruz. 2006;101:517-21. [ Links ]
44. Barlow RE, Gascoyne-Binzi DM, Gillespie SH, Dickens A, Qamer S, Hawkey PM. Comparison of variable number tandem repeat and IS6110-restriction fragment length polymorphism analyses for discrimination of high- and low-copy-number IS6110 Mycobacterium tuberculosis isolates. J Clin Microbiol 2001;39:2453-7. [ Links ]
45. Jou R, Chiang CY, Huang WL. Distribution of the Beijing family genotypes of Mycobacterium tuberculosis in Taiwan. J Clin Microbiol 2005;43:95-100. [ Links ]
46. Lari N, Rindi L, Sola C, Bonanni D, Rastogi N, Tortoli E, et al. Genetic diversity, determined on the basis of katG463 and gyrA95 polymorphisms, Spoligotyping, and IS6110 typing, of Mycobacterium tuberculosis complex isolates from Italy. J Clin Microbiol 2005;43:1617-24. [ Links ]
47. Gomez JE, Leon CI, Inirida M, Leen R, Françoise P. IS6110 fingerprinting of sensitive and resistant strains (1991-1992) of Mycobacterium tuberculosis in Colombia. Mem Inst Oswaldo Cruz 2002;97:1005-8. [ Links ]
48. Cadmus S, Palmer S, Okker M, Dale J, Gover K, Smith N, et al. Molecular analysis of human and bovine tubercle bacilli from a local setting in Nigeria. J Clin Microbiol 2006;44:29-34. [ Links ]
49. Gazouli M, Ikonomopoulos J, Koundourakis A, Bartos M, Pavlik I, Overduin P, et al. Characterization of Mycobacterium tuberculosis complex isolates from Greek patients with sarcoidosis by Spoligotyping. J Clin Microbiol 2005;43:4858-61. [ Links ]
50. Jou R, Chen HY, Chiang CY, Yu MC, Su IJ. Genetic diversity of multidrug-resistant Mycobacterium tuberculosis isolates and identification of 11 novel rpoB alleles in Taiwan. J Clin Microbiol 2005;43:1390-4. [ Links ]
51. Goguet de la Salmoniere YO, Li HM, Torrea G, Bunschoten A, van EJ, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol 1997;35:2210-4. [ Links ]
52. Niobe-Eyangoh SN, Kuaban C, Sorlin P, Cunin P, Thonnon J, Sola C, et al. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J Clin Microbiol 2003;41:2547-53. [ Links ]
53. Zozio T, Allix C, Saribas Z, Alp A, Durmaz R, Fauville-Dufaux M, et al. Genotyping of Mycobacterium tuberculosis clinical isolates in two cities of Turkey: description of a new family of genotypes that is phylogeographically specific for Asia Minor. BMC Microbiol 2005;5:44. [ Links ]
54. Singh UB, Suresh N, Bhanu NV, Arora J, Pant H, Sinha S, et al. Predominant tuberculosis spoligotypes, Delhi, India. Emerg Infect Dis 2004;10:1138-42. [ Links ]
55. Kremer K, van SD, Frothingham R, Haas WH, Hermans PW, Martin C, et al. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J Clin Microbiol 1999;37:2607-18. [ Links ]
56. Gopaul KK, Brown TJ, Gibson AL, Yates MD, Drobniewski FA. Progression toward an improved DNA amplification-based typing technique in the study of Mycobacterium tuberculosis epidemiology. J Clin Microbiol 2006;44:2492-8. [ Links ]
57. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, et al. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variablenumber tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 2006;44:4498-510. [ Links ]
58. Lee AS, Tang LL, Lim IH, Bellamy R, Wong SY. Discrimination of single-copy IS6110 DNA fingerprints of Mycobacterium tuberculosis isolates by high-resolution minisatellitebased typing. J Clin Microbiol 2002;40:657-9. [ Links ]
59. Zanini MS, Moreira EC, Lopes MT, Oliveira RS, Leao SC, Fioravanti RL, et al. Mycobacterium bovis: polymerase chain reaction identification in bovine lymphonode biopsies and genotyping in isolates from Southeast Brazil by spolygotyping and restriction fragment length polymorphism. Mem Inst Oswaldo Cruz 2001;96:809-13. [ Links ]
60. Murray M, Alland D. Methodological problems in the molecular epidemiology of tuberculosis. Am J Epidemiol 2002;155:565-71. [ Links ]
61. Sarmiento OL, Weigle KA, Alexander J, Weber DJ, Miller WC. Assessment by meta-analysis of PCR for diagnosis of smear-negative pulmonary tuberculosis. J Clin Microbiol 2003;41:3233-40. [ Links ]
62. National TB Controllers Association/CDC Advisory Group on Tuberculosis Genotyping. Guide to the Application of Genotyping to Tuberculosis Prevention and Control. Atlanta, GA: US Department of Health and Human Services, CDC; 2004. [ Links ]
63. Brudey K, Gutierrez MC, Vincent V, Parsons LM, Salfinger M, Rastogi N, et al. Mycobacterium africanum genotyping using novel spacer oligonucleotides in the direct repeat locus. J Clin Microbiol 2004;42:5053-7. [ Links ]
64. McNabb SJ, Kammerer JS, Hickey AC, Braden CR, Shang N, Rosenblum LS, et al. Added epidemiologic value to tuberculosis prevention and control of the investigation of clustered genotypes of Mycobacterium tuberculosis isolates. Am J Epidemiol 2004;160:589-97. [ Links ]














