Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista de Salud Pública
Print version ISSN 0124-0064
Rev. salud pública vol.14 no.2 Bogotá Mar./Apr. 2012
1 Molecular Biology Laboratory, Research Center for Endocrine-Metabolic Diseases, Faculty of Medicine, University of Zulia, Maracaibo, Zulia state, Venezuela narraiz1@yahoo.com, narraiz1@gmail.com, vbermudez@hotmail.com, endronix_20@hotmail.com, carem_prieto@hotmail.com, caro_es291085@hotmail.com, anesmu21@hotmail.com
2 School of Nursing, Faculty of Medicine, University of Zulia, Maracaibo, Zulia state, Venezuela burdaneta@cantv.net, rimejia01@cantv.net
3 School of Bioanalysis, Faculty of Medicine, University of Zulia, Maracaibo, Zulia state, Venezuela mpsp85@gmail.com
ABSTRACT
Objective This study was aimed at investigating the frequency of infection by Cp. psittaci and determining its genotype in individuals at potential risk of exposure to the bacteria.
Methodology The study involved 170 individuals: a risk group (n= 96) and a low-risk control group (n=74). Cp. psittaci was detected and genotyped by single-tube nested PCR and ompA gene sequencing.
Results Eight (8.3 %) positive cases were detected in the risk group and 1 (1.4 %) in the control group (p<0.04). Cp. psittaci was found in 16.7 % of pigeons’ fecal samples. Cp. psittaci infection with was more frequent in symptomatic (17.7 %) than asymptomatic (6.3 %) individuals in the risk group. Analysing the genomes isolated from human and bird specimens revealed the presence of genotype B.
Conclusion The presence of Cp. psittaci genotype B in the population being evaluated could have been attributed to zoonotic transmission from pigeons to humans, an underestimated potential public health problem in Venezuela requiring the health authorities’ involvement.
Key Words: Chlamydophila psittaci, Columba livia, ompA, zoonotic transmission, nested PCR (source: MeSH, NLM).
RESUMEN
Objetivo El objetivo de este estudio fue investigar la frecuencia de infecciones por Cp. psittaci y determinar su genotipo en individuos con potencial riesgo de exposición a la bacteria.
Metodología Se incluyeron 170 individuos, un grupo de riesgo (n=96) y un grupo control (n=74). La detección y genotipificación de Cp. psittaci se llevó a cabo por PCR anidada y secuenciación del gen ompA.
Resultados Se detectaron ocho (8,3 %) casos positivos en el grupo de riesgo y 1 (1,35 %) en el grupo control (p<0,04). Cp. psittaci fue detectada en 16,7 % muestras fecales de palomas. En el grupo de riesgo, la frecuencia de infección por Cp. psittaci fue 17,7 % en individuos sintomáticos y 6,3% en asintomáticos. El análisis de los genomas aislados de muestras humanas y aves, revelaron la presencia del genotipo B.
Conclusión La presencia de Cp. psittaci genotipo B en la población evaluada podría ser atribuida a transmisión zoonótica de palomas a humanos, un potencial problema de salud pública en nuestra región que requiere la intervención de autoridades sanitarias.
Palabras Clave: Chlamydophila psittaci, Columba livia, ompA, transmisión zoonótica, PCR anidado (fuente: DeCS, BIREME).
The Chlamydiaceae family consists of two genera: Chlamydia (C) and Chlamydophila (Cp) (1); they include species causing infection in humans and animals (2,3). Chlamydia trachomatis causes urogenital tract infections and is the causal agent of trachoma (3). Chlamydophila pneumoniae causes respiratory infection and seems to be involved in cardiovascular disease, atherosclerosis and neurodegenerative syndromes (4,5).
Chlamydophila psittaci is considered primarily to be a pathogen of veterinary interest; however, several countries are actively investigating this organism’s zoonotic transmission which is responsible for outbreaks of infections (psittacosis) (3,5-8). At least 9 Cp. psittaci genotypes are currently known, based on sequence difference in variable domains of an ompA gene conserved region and it is known that some genotypes are associated with specific bird groups (9).
Human infection can occur by inhaling dried bird faeces or direct contact with infected birds and can display a wide spectrum of clinical manifestations ranging from pneumonia, systemic disease (9,10) to mild or asymptomatic symptoms almost indistinguishable from the common cold (10-14).
Cp. psittaci infection diagnosis has been limited because of a lack of sensitive and specific methods. Culturing Chlamydia is difficult and time-consuming because this group of eubacteria is characterized by an intracellular, biphasic developmental cycle; they are thus reserved for specialized laboratories. Serological tests do not differentiate between Chlamydia species, meaning that techniques involving nucleic acid amplification by polymerase chain reaction (PCR) represent a valuable tool for a definitive diagnosis of psittacosis (3,10,15-17).
As part of a strategy for developing large-scale epidemiological studies in urban areas of Zulia state, this study was aimed at using PCR to investigate Cp. psittaci infection frequency and determine the genotype in individuals having a high risk of exposure to the bacteria due to the massive proliferation of pigeons (Columba livia) in residential buildings’ windows and air conditioner installations.
Population and samples
A cross-sectional study was conducted in north-east Maracaibo, Zulia state, where a massive proliferation of pigeons had been observed in the windows of buildings in residential areas; 105 buildings were selected and residents were invited to participate in the study and provide information through a questionnaire, after signing an informed consent form. The research protocol followed international ethical guidelines and was approved by the University of Zulia’s Council for Scientific and Humanistic Development; the Declaration of Helsinki’s guidelines were followed (revised in October 2008). The questionnaire was designed to collect information related to pet ownership, pet types, occupation, smoking, clinical signs (particularly respiratory ones), medication use, allergies, the presence of birds (pigeons in the windows and air conditioners and the reproductive activity of the pigeons at such sites).
Ninety-six people (56 men and 40 women aged 17-58 years) from families who reported permanent pigeon presence and activity in their homes’ windows and air conditioners agreed to participate in the study. The control group included a sample of 74 individuals (43 men and 31 women aged 19-54 years) whose homes had not been colonized by pigeons. Participants (risk group) were invited to collect stool samples from the niches occupied by pigeons; however, only 78 agreed to collect samples.
The sample consisted of a throat swab taken from fasting participants; the swabs were kept in transport medium (0.2M sucrose, 0.015M Na2HPO4, 0.01M NaH2PO4) and taken to a molecular biology laboratory. Participants were provided with plastic containers for collecting pigeons’ stools.
DNA extraction
A previously-described procedure for C. trachomatis research was used for DNA extraction (18); 5 µl of the DNA sample was used for amplification assays.
The lysis procedure described above was followed for extracting DNA from faecal samples (0.5 to 0.7 g) after debris had been removed by 3 washes with 1.5 ml phosphate buffer saline (0.1M Na2HPO4, 0.1M NaH2PO4, pH 7.5); 10 µ L of sample was used for amplification assays.
Chlamydophila psittaci detection by PCR
A previously-described single-tube nested PCR technique was used, with some modifications (16). The mix included two pairs of oligonucleotides, a set of family-specific primers targeting Chlamydiaceae family members’ (Cp pneumoniae, Cp. psittaci and C. trachomatis) conserved 16S RNA ribosomal gene (16S rDNA) sequences (436 bp) and a second set of primers targeting Cp. psittaci species-specific variable regions (126 bp) binding to the family-specific PCR product’s internal sequences. All assays were carried out simultaneously with the corresponding positive and negative controls, using the same reagent mixture.
The reaction mixture (50 µl final volume) consisted of 10 µl Taq DNA polymerase buffer 5X (Promega), 1.5 mM MgCl2, 200 µM of each deoxyribonucleotide, 20 pmol of each primer, 1.25 U Taq DNA polymerase and 200 ng DNA. Amplification consisted of an initial denaturing at 96ºC for 5 min, 10 cycles of 94°C for 1 min, 50°C for 1 min and 72ºC for 1 min, followed by 35 cycles of 94°C for 1 min, 55°C for 1 min and 72ºC for 1 min. The final extension step at 72°C lasted 8 minutes. PCR products were analysed by electrophoresis on 2.5% agarose gels, stained with ethidium bromide (5 µg/ml), visualised by UV lamp and photographed with UVP (Digi Doc, USA) photo documentation system. G210A PCR DNA ladder (Promega) was used as molecular weight marker.
Genotyping Chlamydophila psittaci
Cp. psittaci was genotyped in both human and bird samples by sequencing ompA gene’s variable domain (VD) region using primers and the previously-described procedure (9). PCR products (1,041 bp) were purified from the gel using Wizard Gel and PCR Clean-Up System (Promega), amplified by 25 cycles and sequenced in both directions using ABI Prism 310 automatic DNA sequencer (PE Applied Biosystem, Foster City, CA) with a BigDye v3.0 kit (Applied Biosystems). The results were analyzed using ABI Sequence Navigator software (v.3.1) and compared by alignment with ompA gene sequences from the following C. psittaci genotypes available in GenBank: A (AY762608), B (AY762609), D (AY762610;) E (AY762611), F (AY762612), E/B (AY762613); the Blast algorithm was used (http://www.ncbi.nlm.nih.gov).
Statistical analysis
The results were analyzed using SPSS software (version 12 for Windows). Descriptive analysis gave frequencies for each variable, along with their respective percentages. Possible association between variables was estimated by comparing observed and expected values through contingency tables, the Chi2 test and Pearson’s correlation; a p<0.05 was considered statistically significant.
Massive colonization by pigeons in north-east Maracaibo, Zulia state, is invading residential buildings. Since there is no health control strategy monitoring such colonization, these buildings have become contaminated with pigeon faeces, setting inhabitants of the urban areas involved at risk. This study thus detected and genotyped Cp. psittaci in both human respiratory samples and pigeon stools.
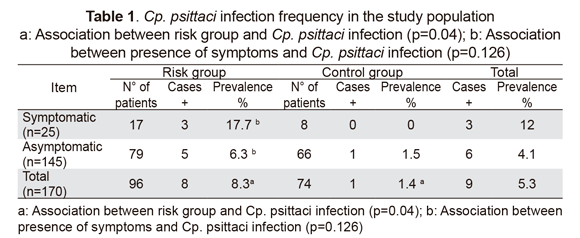
The single-tube nested PCR strategy used in this study led to amplifying a variable region within the 16S rDNA gene sequence that is specific to Cp. psittaci. Figure 1 gives an example of PCR amplification of 12 samples, revealing a 126 bp fragment in some lanes and indicating the presence of the Cp. psittaci genome. Eight positive cases were found in the risk group and 1 in the control group (Table 1), leading to estimating pathogen 8.3% and 1.4% prevalence, respectively (Chi2=4.063; p=0.04).
Only 3/8 (37.5 %) of risk group individuals for which Cp. psittaci infection was demonstrated had respiratory symptoms (Table 1), consisting of cough, pharyngitis and rhinitis during the three weeks prior to test whilst the other five cases (62.5 %) exhibited no symptoms at the time of this study. Even when Cp. psittaci infection prevalence in the symptomatic subgroup (17.7 %) was higher than in the asymptomatic one (6.3 %), no statistically significant difference was found between subgroups (Chi2=2.346; p=0.126).
The questionnaire revealed that the target population had no occupational risk associated with handling birds or having pet birds, but reported high pigeon presence and activity in buildings’ windows and air conditioning, particularly in respondents’ bedrooms.
Analyzing the ompA gene sequence amplified from the genomes isolated from human respiratory samples (7 samples: 6 risk group and 1 control group) and 9 of the 13 stool samples from pigeons revealed 100% identity with reference sequence genotype B (GenBank Accession number: AY762609).
A major risk factor which may contribute to Cp. psittaci zoonotic transmission concerns the fact that when pigeons increase their egg-laying rate there is a concomitant increase in bacterial excretion during egg incubation and chick feeding (12,19,20). This is particularly serious considering that residences’ windows and air conditioning spaces, especially bedrooms, are used by pigeons during breeding and feeding.
Heddema et al., 2006 (19) reported that 5 %-10 % of pigeons’ faecal samples in urban populations in Amsterdam were infected with Cp. psittaci, although this work did not show zoonotic transmission events. However, the bacteria was detected directly in exposed populations’ clinical samples and faecal samples from birds collected by participants in this study which could have represented direct evidence of zoonotic transmission from birds to humans.
Given that conventional serological, PCR-RFLP and real-time PCR techniques do not allow accurate discrimination of currently-known genotypes (21,22), the Cp. psittaci genotype was determined in this study by ompA gene sequencing. The B genotype being detected in the samples was consistent with other studies indicating that this genotype has been predominantly isolated in columbiforme birds (9,23,24). The genotype could not be obtained in all samples due to failures in ompA gene fragment amplification. This may have occurred because this gene is represented by a single copy per genome, unlike ribosomal RNA genes.
These results may partially explain an asymptomatic course of infection in some individuals, because it has been reported that Cp. psittaci genotypes A, D and E/B cause severe respiratory symptoms, while genotype B has been isolated from birds and humans showing mild clinical manifestations or having asymptomatic infection (9,25,26).
This does not rule out the possibility that these individuals may develop symptoms later on and all genotypes should be considered potentially pathogenic for humans (3,20).
Another explanation is that these individuals may have developed protection against severe infection due to chronic exposure to the infectious agent, as suggested by other authors (24).
The risk population reported not having pet birds in their homes and had no occupational risk related to handling birds, suggesting a direct relationship between Cp. psittaci infection and pigeon colonisation in their buildings. Instead, Cp. psittaci infection was only detected in one case (1.4%) in the non-exposed population (control group).
The single-tube nested PCR strategy used in this study met specificity criteria because it could amplify a variable region within the 16S rDNA gene sequence specific for Cp. psittaci and there was higher sensitivity as a result of increased target DNA availability for amplification during the first cycles and species-specific product enrichment. The strategy used in this study may be implemented as a routine assay to help diagnosis and epidemiological characterisation of Cp. psittaci transmission in humans and animals.
This study, the first in our region, has made it evident that there was significant Cp. psittaci genotype B infection in the urban population being studied. This could have been attributed to zoonotic transmission from birds to humans, particularly pigeons, which have become important reservoirs of Cp. psittaci, an underestimated potential public health problem in our region that requires the involvement of health authorities.
Author contributions: NA, VB, BU, MPS, CP, CE, AM, EM conceived and designed the experiments, NA, MPS, CP performed the experiments, NA, VB, AM EM analysed the data, NA, AM, EM contributed reagents/materials/analysis tools, NA, EM wrote the paper and VB, BU, MPS, CP, CE, AM, EM revised the paper.
Acknowledgment: This study was supported by the Council on Scientific, Humanistic and Technological Development, University of Zulia (CONDES-LUZ), through grant CC-0736-08, Maracaibo, Zulia, Venezuela.
1. Everett KD, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999; 49:415-440. [ Links ]
2. Corsaro D, Venditti D. Emerging chlamydial infection. Crit Rev Microbiol. 2004; 30:75.106. [ Links ]
3. Corsaro D, Greub G. Pathogenic potential of novel Chlamydiae and diagnostic approaches to infections due to these obligate intracellular bacteria. Clin Microbiol Rev. 2006; 19:283-297. [ Links ]
4. Stratton CW, Sriram S. Association of Chlamydia pneumoniae with central nervous system disease. Microb Infect. 2003; 5: 1249-1253. [ Links ]
5. Bedson SP, Western GT, Simpson SL. Observations on the aetiology of psittacosis. Lancet. 1930; i:235-236. [ Links ]
6. Pannekoek Y, Visser C, Duim B, Heddema ER. Chlamydophila psittaci infections in The Netherlands. Drugs Today. 2009; 45 Suppl B:151-157. [ Links ]
7. Verminnen K, Vanrompay D. Chlamydophila psittaci infections in turkeys: overview of economic and zoonotic importance and vaccine development. Drugs Today. 2009; 45 Suppl B:147-150 [ Links ]
8. Iijima Y, Akiyoshi K, Tanaka S, Nukina M, Ito M, Haruta T, Inoue A, Ando S, Kishimoto T. Psittacosis outbreak at an avian exhibition. Kansenshogaku Zasshi. 2009; 83:500-505. [ Links ]
9. Heddema E, van Hannen E, Duim B, Vandenbroucke-Grauls CMJE, Pannekoek Y. Genotyping of Chlamydophila psittaci in Human Samples. Emerg Infect Dis. 2006; 12: 1989-1990. [ Links ]
10. Stewardson AJ, Grayson ML. Psittacosis. Infect Dis Clin North Am. 2010; 24: 7-25. [ Links ]
11. Harkinezhad T, Verminnen K, De Buyzere M, Rietzschel E, Bekaert S, Vanrompay D. Prevalence of Chlamydophila psittaci infections in a human population in contact with domestic and companion birds. J Med Microbiol. 2009; 58:1207-1212. [ Links ]
12. Haag-Wackernagel D, Moch H. Health hazards posed by feral pigeons. J Infect. 2004; 48:307-313. [ Links ]
13. Verminnen K, Duquenne B, De Keukeleire D, Duim B, Pannekoek Y, Braeckman L, Vanrompay D. Evaluation of a Chlamydophila psittaci infection diagnostic plattform for zoonotic risk assessment. J Clin Microbiol. 2008; 46:281-285. [ Links ]
14. Petrovay F, Balla E. Two fatal cases of psittacosis caused by Chlamydophila psittaci. J Med Microbiol. 2008; 57:1296-1298. [ Links ]
15. McElnea CL,Cross GM. Methods of detection of Chlamydia psittaci in domesticated and wild birds. Aust Vet J. 1999; 77:516-521. [ Links ]
16. Messmer TO, Skelton SK, Moroney JF, Daugharty H, Fields BS. Application of a nested, multiplex PCR to psittacosis outbreaks. J Clin Microbiol. 1997; 35:2043-2046. [ Links ]
17. Hewinson RG, Griffiths PC, Bevan BJ, Kirwan SES, Field ME, Woodward MJ, Dawson M. Detection of Chlamydia psittaci DNA in avian clinical samples by polymerase chain reaction. Vet Microbiol. 1997; 54: 155-166. [ Links ]
18. Arraiz N, Marcucci R, Colina S, Reyes F, Rondón N, Bermúdez V, Reyna N. Infección por Chlamydia trachomatis en mujeres consultantes en Maracaibo, Venezuela. Rev Salud Pública (Bogotá). 2008; 10: 615-620. [ Links ]
19. Heddema E, Sluis S, Buys J, Vandenbroucke-Grauls MJ, van Wijnen J, Visser C. Prevalence of Chlamydophila psittaci in Fecal Droppings from Feral Pigeons in Amsterdam, The Netherlands. Applied Environm Microbiol. 2006; 72: 4423-4425. [ Links ]
20. Telfer BL, Moberley SA, Hort KP, Branley JM, Dwyer DF, Muscatello DJ, Correll PK, England J, McAnulty M. Probable psittacosis outbreak linked to wild birds. Emerg Infect Dis. 2005; 11:391-397. [ Links ]
21. Nordentoft S, Kabell S, Pedersen K. Real-Time Detection and Identification of Chlamydophila Species in Veterinary Specimens by Using SYBR Green-Based PCR Assays. Appl Environ Microbiol. 2011; 77:6323-6330. [ Links ]
22. Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp Immunol Microbiol Infect Dis. 2010; 33:473-84. [ Links ]
23. Vanrompay D, Harkinezhad T, van de Walle M, Beeckman D, van Droogenbroeck C, Verminnen K, Leten R, Martel A, Cauwerts K. Chlamydophila psittaci transmission from pet birds to humans. Emerg Infect Dis. 2007; 13(7):1108-10). [ Links ]
24. Beeckman DSA, Vanrompay DCG. Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin Microbiol Infect. 2009; 15: 11-17. [ Links ]
25. Sachse K, Laroucau K, Hotzel H, Schubert E, Ehricht R, Slickers P. Genotyping of Chlamydophila psittaci using a new DNA microarray assay based on sequence analysis of ompA genes. BMC Microbiology. 2008; 8: 63-74. [ Links ]
26. Greco G, Corrente M, Martella V. Detection of Chlamydophila psittaci in Asymptomatic Animals. J Clin Microb. 2005; 43: 5410-5411. [ Links ]













