Fusarium disease is the second reason of fungal infection caused by filamentous fungi, after aspergillosis 1. The incidence of this fungal infection varies according to the region and is more common in regions with warm and humid weathers 2. Fusarium fungi can infect humans through the inhalation of micro particles from the polluted environment. Other less frequent forms of contamination occur through direct contact with skin structures damage, in-hospital contamination of water deposits and nearby construction sites, from which the dispersion of the conidia in micro particles is capable of reach the respiratory tract 1.
In humans some Fusarium species cause infections as onychomycosis, keratitis, endophthalmitis, fungemia by using catheters, peritonitis, skin and subcutaneous infections; and less frequent, osteomyelitis, arthritis, otitis, sinusitis and brain abscess, and invasive fungal infections 3-5.
The clinical form of fusariosis depends largely on the immune status of the host. Superficial and localized diseases occur mostly in immunocompetent patients and invasive and disseminated diseases affect immunocompromised patients 6. Unlike the immunocompetent host, where onychomycosis and keratitis are the most frequent manifestations, disseminated fusariosis is the most common form of presentation in severe immunosuppression states 1. The most commonly associated predisposing factors are: prolonged neutropenia especially in patients with leukemia or transplanted hematopoietic progenitors; therapy with corticosteroids or cytotoxic chemotherapy 7. The typical pattern is a granulocytopenic patient who has received a treatment prolonged with broad-spectrum antibiotics due to fever unknown etiology. A higher incidence has also been observed of infections by these filamentous fungi in patients with solid organ transplantation 7.
The distinctive feature of disseminated fusariosis is the appearance of purpuric skin nodules with an area of central necrosis. In general, the biopsy of these nodules reveals the presence of septate hyaline hyphae with branches that invade the dermal blood vessels. The cultures of the biopsy material and the blood cultures (positive in more than 75%) are useful in the diagnosis of Fusarium infection. Clinically, it manifests with fever and large ulcerated skin lesions that progress to necrosis. This appearance complicates the distinction of this fungus with Aspergillus. However, unlike aspergillosis, patients with disseminated fusariosis usually have positive blood cultures 7. The prognosis is reserved since the mortality of the disseminated form is around 80% 1.
Fusarium houses about 100 species, most reported as human pathogens are F. solani (Mart.) Sacc and F oxysporum Schlechtendal 8-10 and less frequently F verticillioides (Sacc.) Nirenberg 9,10, F proliferatum (Matsush) 11,12, F. dimerum Penz13 and F. sacchari (EJ Butler & Hafiz Khan) W. Gams 14; however, in most clinical cases the species is not reported because it is one of the most heterogeneous and difficult to classify fungi 15. Isolates of F. solani are more resistant to most of the antimicotic available than those of F. oxysporum.
Recently, the use of molecular methods based on sequence analysis of multiple genes has contributed to a more accurate identification of Fusarium and has showed great inter -and intra- species diversity, that in some cases represent complexes of species 16,17. Among the most important molecular markers used to identify Fusarium are the 5' region of the 28 s rDNA subunit 18 and the gene encoding the elongation factor of translation 1α (TEF1-α), considered latter as the most informative gene for identification to species or species complex of the genus 19.
In Colombia, the identification of clinical isolates of Fusarium to the species level is incipient. However, there are studies that suggest Fusarium spp. as the main causative agent of onychomycosis, among non- dermatophyte fungi 20-22; and, cases of invasive fusariosis 1, a single pulmonary nodule in an immunocompetent patient 23 and keratitis 24 has been reported.
Having into account the simplicity of the PCR technique, the objectives of this study were to detect clinical isolates belonging to the genus Fusarium and the species F. oxysporum, F. solani, F. verticillioides and F subglutinans, by PCR with specific primers. Also, to identify the isolates by partial sequencing of the gene 28 s rDNA and the TEF1-α, in order to validate the PCR technique to detect the most common species of Fusarium. This study will contribute to a faster and more accurate diagnosis that is essential for timely and accurate treatment, taking into account the high resistance of Fusarium to most of the antifungal agents available.
MATERIALS AND METHODS
Origin of isolates
101 clinical isolates were obtained from symptomatic individuals, submitted to the laboratory of Medical Mycology and Experimental (MME) of the Corporation for Biological Research (CIB) in the period between 2004 and 2006. Samples were taken from nails and toes, cornea, sinuses, skin, or discharge; they were cultured in Sabouraud Agar, PDA and Mycosel media and incubated at room temperature for a period of seven to 14 days.
Preliminary identification of Fusarium
The initial identification of isolates of Fusarium spp. was performed by observation at the microscopic of plate with lacto phenol blue, direct examination with 20% KOH Chinese ink and by description of the macroscopic features of colonies after a period of incubation of eight to ten days at 23 °c. Pure colonies identified as Fusarium spp. were stored in vials with sterile water.
DNA extraction from monosporic culture
The monosporic culture of each isolate of Fusarium spp. was carried out by Giraldo D. in 2010 25. A portion of mycelium (approx. 100 mg) was taken and it was macerated with liquid nitrogen and equal volume of lysis buffer (1 mM EDTA, 10 mM Tris HCL, pH 8, sodium acetate was added 2M, 100 mM NaCl, Triton X - 100 2% and SDS 1%), for DNA extraction 26.
Detection by PCR of Fusarium to the genus and species level
PCR detection of isolates belonging to the genus Fusarium was performed with the primer pair Fus1/Fus2, which have as target the 28S rDNA subunit 27. Detection of F oxysporum was performed with the primer pair 0x31/ 0x32 28. F solani isolates were detected with the primer pair Fusorev and Fusofor, which have as target the 18S rDNA subunit 29. The previous thermocycler conditions used were those reported 25,27-29.
Specificity of the primers used in the detection of the genus Fusarium and the species F. oxysporum and F. solani
The specificity of the primers used for the detection of Fusarium and F. oxysporum and F. solani species was assessed by PCR with DNA of: Acremonium sp., Alternaria sp, Aspergillus flavus, A. fumigatus, A. niger, A. terreus, A. versicolor, Candida albicans, C. tropicalis, Malassezia furfur, Neoscytalidium dimidiatum, Penicillium sp., Scedosporium apiospermun and Trichosporon asahii, fungi commonly diagnosed in the Medical Mycology Unit of the CIB.
Partial sequencing of 28 s rDNA
Partial sequencing of the 28S rDNA with the primer pair Fus1/Fus2 was performed for 34 isolates. The selection of the isolates was carried out as follows: 10 isolates identified as F. oxysporum with 0x31/0x32 primers; five isolates, identified as F. solani with the primers Fusorev/Fusofor; and 16 isolates, whose species could not be detected with any of the primer pairs 0x31/0x32, Fusofor/Fusorev. Additionally, three isolates (31 791, 32 892 and 32 989) from the collection the Universidad de los Andes Laboratory of Microbiology and Plant Pathology (LAMFU) were included. The PCR products were sent to Macrogen (South Korea) for sequencing in both directions, in an ABI Prism 3730XL.
The sequences were edited with Genious Pro software (version 4.5 Biomatter Inc). A multiple alignment was performed with Clustal W using the consensus sequence of each isolate; it included five sequences of the 28S rDNA of different Fusarium species, reported in the GenBank database (accession number FJ614650.1F, AY097318.1, AY097316.1, DQ236682.1 and EU926284.1, http://www.ncbi.nih.gov/GenBank/). A phylogenetic tree was constructed by Neighbor-Joining, with 1 000 replicates and a supported Bootstrap above 50%, by the Jukes-Cantor model with the option "pairwise deletion" with the MEGA 6 software. In addition, each of the consensus sequences was identified by BLAST (www.ncbi.nlm.nih.gov/blast/) analysis to confirm their identity.
Partial sequencing of TEF1-α
41 isolates selected from the results obtained by detection and sequencing of the 28S rDNA gene, were also analyzed by partial sequencing of TEF1-α gene. The PCR amplification was done with the EF1-EF2 primers 30, in a thermocycler T3000 (Biometra), with the following parameters: initial denaturation at 94 °C for five min., followed by 35 cycles of 95 °C for 30 s., 57 °C for one min., 72 °C for one min., and a final extension at 72 °C for seven min. The amplified products were purified with the QIAquick PCR Purification kit (QIAGEN, Germany). Sequencing was done in both directions with the primer pair EF3 and EF22T 31, with the Big Dye Terminator Cycle methodology (Applied Biosystems, USA), in an ABI Prism 3730XL sequencer of Macrogen (South Korea).
The species complex (hereafter the species) of each isolated was determined with the consensus sequence by BLAST on Fusarium ID database (http://www.fusa-riumdb.org/, table 1). With all sequences, and eight sequences of Fusarium ID database and eight sequences of the GenBank; a multiple alignment was performed using the algorithm Clustal W. A phylogenetic tree was constructed by Neighbor-Joining, with 1 000 replicates and a supported Bootstrap above 50%, by the Kimura-2 parameter model with the option "pairwise deletion" with the MEGA 6 software.
Table 1 Molecular identification of Fusarium isolates obtained from clinical samples

* Identification obtained by comparison to the GenBank database using the BLAST tool. ** Identification obtained by comparison to Fusarium ID database using the BLAST tool. ₪ Unamplified strains with any of the primers
Electrophoresis
All PCR-amplified fragments were observed on agarose gel electrophoresis in 1,5% (w/v), stained with ethidium bromide (0.5 µl/10 ml), the size of the fragments was verified by comparison with the molecular weight marker 100 bp GeneRuler Plus (Fermentas).
RESULTS
Preliminary identification of Fusarium isolates
101 clinical isolates of Fusarium were studied in this research. Macroscopically, most colonies were characterized by their cottony aspect, salmon pigmentation, purple, lilac, light brown, yellow or gray. Microscopically with hyaline hyphae, septate and filamentous; septate macroconidia, fusiform with characteristic appearance of alantoespora, oval microconidia and in some cases chlamydoconidia of thick wall 25.
PCR detection of clinical isolates of Fusarium
PCR detection of Fusarium isolates with the primers Fus1/Fus2 allowed to obtain PCR products of the expected size for all 101 isolates. The primers showed that they are specific to the genus Fusarium, since they did not amplify any fragment from the DNA of all the other fungus genera evaluated.
PCR detection of F. oxysporum and F. solani isolates with the primer pairs Fusofor/Fusorev and 0x31/0x31, respectively, revealed the expected fragment from 52 (51.4%) and 28 (27,7%) out of 101 isolates, respectively.
The specificity of these two pairs of primers showed no amplification of any DNA fragment from the other genera of fungi evaluated.
Partial sequencing of the 28s rDNA gene and TEF1-α
A total of 34 and 41 isolates were analyzed by partial sequencing of the 28s rDNA and TEF1-α genes, respectively. Besides 10 isolates being detected by PCR as F oxysporum, the rDNA also allowed to identify them as F oxysporum; likewise, TEF1-α sequencing confirmed the identity of all as F. oxysporum, except for the isolate 58 025, which was identified as F solani. In the dendrogram constructed from rDNA sequences, the 58 025 was allocated in the cluster I, with 100% of identity with strain ATCC MYA- 3970 of F oxysporum (Figure 1), but in the dendrogram constructed with sequences obtained from TEF-α, it was located in one of the cluster of F solani (Figure 2).
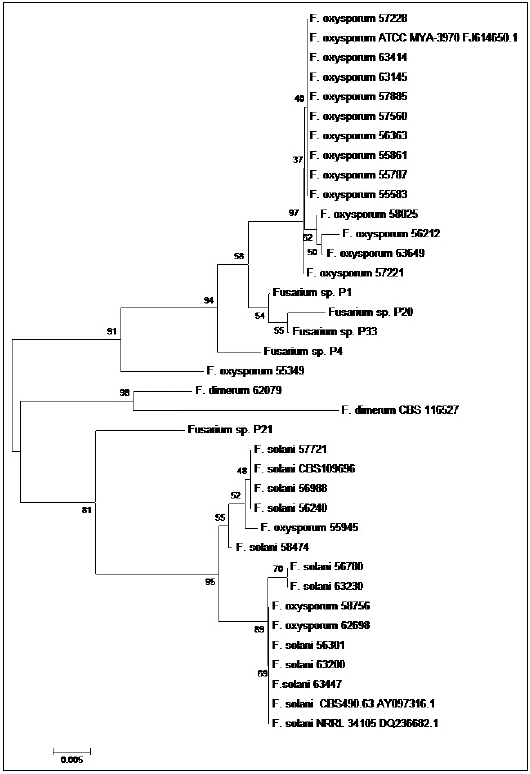
Figure 1 Dendrogram of the 28S rDNA region of 34 clinical isolates of Fusarium spp., and five sequences of Fusarium spp., taken from the GenBank, constructed by Neighbor Joining with Bootstrap support > 50%, with 1000 replications

Figure 2 Dendrogram of the TEF1-α region of 41 clinical isolates of Fusarium spp, eight sequences of Fusarium spp., taken from the GenBank and eight sequences from Fusarium ID, constructed by Neighbor-Joining, waith supporting Bootstrap, 1000 replications, under the model of Jukes-Cantor
Similarly, of five isolates detected by PCR as F. solani, rDNA sequencing indicated its identity as F. solani; the TEF 1-α gene confirmed the identity of the isolates as F. solani, but not to 55 945 which was identified within the complex F. oxysporum. In the dendrogram constructed by rDNA sequences such isolate was placed inside the F. solani group, close related to the F. solani CBS109696 strain (Figure 1); contrarily, in the dendrogram constructed with the TEF1-α sequences, this isolate was placed with 100% identity with the F. oxysporum strains, NRRL38515 and MYC163 -05 (Figure 2).
Out of 21 isolates not detected by PCR with any of the two pairs of primers used to detect F. oxysporum, F. solani, eight were analyzed by partial sequencing of the genes 28s rDNA and/or TEF1-α. Of those isolates, three were identified as F. oxysporum and four as F. solani by the two genes. The isolate 56 665 was only identified as Fusarium spp., for both genes. The isolate 62 698, not detected by pcR with any of the four primer pairs, was identified by sequencing of the gene 28s rDNA as F. solani, with 100% of identity with strain CBS 490.63 of F solani, and by the TEF1-α gene - as F. oxysporum, with 100% of identity with the sequences of strains NRRL38515 and MYC163 -05 of F. oxysporum (Figures 1 and 2).
DISCUSSION
Fusarium disease is the second reason of fungal infection caused by filamentous fungi, after aspergillosis, mainly affecting immunocompromised patients. Clinical presentation depends on the route of entry of the fungus, the intensity and duration of immunosuppression. The prognosis is reserved since the mortality of the disseminated form is around 80% 1,7.
In recent years, there has been a remarkable increase of reports of these fungal infections, possibly, due to the alteration of the bacterial flora product of the overuse of topical antibiotics, the extensive use of corticosteroids and immunosuppressive drugs, as well as the improvement of diagnostic methods 7. Climate changes are predisposed to this type of fungal diseases 7.
About 15 species of Fusarium have been reported as causative agents of human diseases, among the most common are F. solani and F. oxysporum.7,14,21,32,33. In Colombia, the identity of Fusarium isolates to the species level is rarely reported, although the cases are in rise. Between January 1980 and September 1989, 87 cases of dermal and ocular Fusarium infection were diagnosed in the CIB 20 and over 2003 and 2004 a total of 128 isolates from 137 patients with onychomycosis caused by Fusarium spp. were reported in the Laboratorio Especializado de Micologiía Médica (LEMM) in Bogotá 22.
Identification of the species level of fungi belonging to Fusarium using classical methods is difficult. It is time consuming and it has limitations in terms of sensitivity and specificity 34,35. On the other hand, Fusarium taxonomy complexity is widely known 18,31,36-41.
Molecular diagnostic methods allow faster and higher resolution and sensitivity. The correct and rapid diagnosis of the agent involved is essential for the implementation of appropriate treatment, which if done correctly, helps to reduce the high mortality rates of high-risk patients 34.
This study validates the use of two pair of PCR primers for fast and precise detection of Fusarium isolates belonging to F. oxysporum or F. solani, which represents more than 90% of the isolates involved in human fusariosis 21,22,34,42-44. In our work, F. oxysporum was the most common specie, followed by F. solani. These results are in agreement with those found in another city of Colombia, Medellin, by partial sequencing of TEF1-a and rpb2 genes, 45 although some other works report F. solani as the most common specie 21,22,46.
Partial sequencing of the 28 s rDNA gene has suggested the usefulness of the primers 0x31/0x32 and Fusorfor/Fusorev to detect isolates of F. oxysporum and F. solani. Several authors report the ribosomal regions as uninforma-tive for differentiation of Fusarium species 11, however this study highlights its usefulness in detecting the genus and the species complex F. oxysporum and F. solani, in concordance with other reports 44,47,48.
The TEFF-α sequence, considered the best molecular marker to identify Fusarium isolates 31, also confirmed the detection of the PCR primers 0x31/0x32 and Fusorfor/Fusorev F. oxysporum; besides, as expected, the TEFF-α sequences identified 16 isolates that were not detected with any of the specific primers.
The isolate 56 665 identified only as Fusarium spp., both by sequencing of the 28 s rDNA and by the TEF1-α gene, should be considered in a subsequent study