Introduction
One of the most studied areas that is in constant development is medicine, it is due to the existence of a great variety of different diseases that affect human health. These illnesses can be as evident as the flu, which can be treated at home without any kind of complications, or they can go unnoticed until the symptoms become obvious, at which point when the lack of timely treatment can generate some disability for the patients or even be fatal. That is the most important reason for the importance of early diagnosis, because there are illnesses whose progression can be stopped or controlled even if there is no cure, all with the help of rigorous controls to identify injuries, signs, symptoms, organism performance changes, and so on [1].
While it is true that these diseases could be found at a first check by a professional, it is also true that they can go unnoticed because their characteristics at the early stages used to be subtle and almost invisible, until their progress began to be evident, as it happens with Alzheimer's, diabetes, cancer, etc. One of the most studied illnesses, due to its high mortality rate and incidence, is cancer. In the beginning, it can be in any part of the human body, and in some situations, its existence is unknown until that moment when the tumor starts to grow uncontrolled or spread to other parts of the human body. One of the most aggressive types of cancer is known as skin malignant melanoma. First, it looks like a lesion like a mole that, depending on certain features, can be detected, and studied in time to avoid its spreading (also known as metastasis) and cure it by decreasing its mortality risk. [2]
Usually, the first diagnostic given by a professional to determine if a skin mole is not a malignant melanoma is a visual technique known as the ABCDE rule; it is used to check different physical characteristics of the mole. However, as it is a visual technique, it can generate false positives, and it is believed the sensibility of a clinical diagnosis made by a professional in medicine varies between 70 and 88 %, while in the case of a dermatologist with experience, it is estimated to be greater than 82 % [3] [4]. Due to this percentage and the existence of false positives, this paper proposes to develop a system based on artificial intelligence and computer vision by constructing a convolutional neural network capable of learning by itself, from a frame of images taken from ISIC'S web page [5], how to determine if the lesion is malignant or not, with the goal of outperforming medical professionals, avoiding the need for a possible biopsy, and making it easier to obtain an early dialysis.
Through the past few years, artificial intelligence (AI) has become one of the most widely used technologies thanks to its great variety of applications, versatility, and adaptability to any kind of process that can be automated, controlled, and learned, all in the hope of being to be able to optimize duty time and get more accurate results than those generated by human hand work. The present paper shows the development of a convolutional neural network designed to identify melanomas by getting a prediction at its output, which means the lesion tested is malignant or benign; thus, the accuracy percentage from this classifying method is just a little higher than the approximate given by an experienced dermatologist [3], providing up to 90 % of benign lesions successfully. The second stage consists of the convolutional layers, which are all followed by 2x2 pooling layers to extract and filter only the most relevant characteristics of every image. Finally, the third stage corresponds to the fully connected layer where the classification takes place, where the given result represents the prediction given by the network if the lesion is or is not malignant.
Following the idea exposed in the previous paragraphs, the objective of this work is to develop a system to analyze images of skin lesions via CNN to obtain a model capable of detecting malignant skin tumors of the melanoma type in humans. This is especially true considering that melanoma is one of the most aggressive types of skin cancer due to its tendency to metastasize and the fact that it closely resembles benign skin lesions, which makes it hard to diagnose and makes it mandatory to perform several unnecessary biopsies.
Previous studies
Currently, some ways to study or special examinations used by dermatologists to diagnose melanoma are based on visual patterns, and they are needed to get an early diagnosis and timely treatment if needed. Some of those special treatments are:
The naked eye study and the methodology of the Glasgow 7 points checklist are presented in [3] and [2] in Table 1 to assess suspicious skin lesions. Apply it as a triage means to evaluate the features in Table 1 and if the score obtained is 3 or more, then it is recommended to refer to a specialist via an urgent suspected cancer pathway [2].
Table 1 Weighted Glasgow 7-point checklist. [2]
Source: Jones, Owain T [2].
The "ABCDE" rule is another kind of naked eye assessment based on asymmetry, border, color, diameter, and evolution examination, where changes on any of the mentioned features point to suspicious melanocytic lesions, increasing the possibility of the existence of malignant melanoma, making referral to the dermatologist necessary [2], [3].
A dermoscopic pattern classification based on color pigmentation [6] is used to detect cutaneous metastases in malignant melanoma cases, classifying based on four color patterns: mixed, brown, pink, and blue.
On the other hand, as technology has advanced and become more versatile, some research projects devoted to melanoma classification have emerged, with a focus on automated processes. To increase efficiency in any medical diagnostic process, such as skin cancer cases, the world has begun to work with some computational strategies, such as in [7], where the Raman spectra are going to be preprocessed for features extraction and recently evaluated by a feedforward neural network for automatic classification of skin cancer, or in [8], where the authors present a technique based on histogram averaging. Other techniques studied in the past years are based on dermoscopic or non dermosco-pic images, taking from them some specific features (like contour, color, shape, etc.) by the use of different processing tools, and finally getting the classification expected, as shown in [9], [10], [11] and [12].
Recently, works related to images have started to favor the use of Convolutional Neural Networks (CNNS) as the deep learning method to automate procedures, and melanoma classification is not the exception, as presented in [13], [14], [15], [16], [17], [18] and [19], where CNNS are used together with other techniques to get better results: A very deep fully convolutional residual network (FCRN) with more than 50 layers is employed for accurate skin segmentation, and its capability is further improved by incorporating a multi-scale contextual information integration scheme, to cope with degeneration and overfitting problems when the network goes deeper. Based on segmentation results, a very deep residual network (DRN) is employed to be able to distinguish melanomas from non-melanoma lesions more precisely [13]. A 19-layer CNN with 290 129 trainable parameters and a stride fix of 1 uses ReLU as the activation function and at the output, a sigmoid activation function, was trained end to end together with a novel loss function that handles the lesion background imbalance of pixel-wise classification for image segmentation based on Jaccard distance [14].
In [15], they affirm that segmentation of the lesion in the skin image is a crucial step in the process for most detection algorithms. The first key to obtaining high accuracy in subsequent steps in the process is to apply accurate segmentation. They propose a methodology that works by combining automated segmentation and the CNN module. For training, 900 images from ISIC 2016 were used. These images were also classified into three types based on their characteristics: Melanoma, Seborrheic keratosis, and Nevus. Images were segmented by a technique called Generalized Gaussian Distribution (GGD), and then CNN was applied for feature extraction and posterior classification.
The two-stage methodology was proposed in [16]. First, the training images were preprocessed by excluding a specific range of gradients to discard the illumination effects. A K- means classifier was then applied to produce a segmentation mask that was used to extract the lesion's region and a Gaussian filter is finally applied to minimize normal skin texture's effects on the classification process. In the second stage, the architecture of the CNN proposed was built with two convolutional layers with a 5x5 kernel, all stacked together with pooling layers after each convolution, getting at the output layer 2 categories (Melanoma and Nevus).
Other works have developed techniques such as the Fisher Vector (FV) and the Support Vector Machine (SVM) [17], where instead of adopting preprocessing techniques such as lesion detection, segmentation, and training the entire convolutional neural network, the proposed scheme consists in a combination of the advantages of both deep features and local descriptors, encoding methods efficiently by proposing a hybrid framework, which combines feature extraction via CNN from multiple sources with feature extraction via CNN from multiple sources.
In [20], Jorge Alexander Angeles Rojas proposes a model composed of four convolution blocks to perform characteristic extraction and then the classifier in the final layer where the LI-SVM loss function is implemented, resulting in a hybrid model of a convolutional neural network and a support vector machine (CNN+SVM) to classify BCC (basal cell carcinoma), with an f1-score of up to 96.2 %.
More recent studies have developed various CNN-based techniques, such as Sahar Nisir Haghighi in [21], where data augmentation was used to compensate for the lack of training data required to feed the proposed method, which merges features between the CNN architecture and the Support Vector Machine classifier, achieving a promising accuracy of 89.52 %; and in [22], a machine learning-based technique using CNN is proposed to classify 7 types of skin diseases, in this case, along with the CNN, transfer learning used to improve the classification accuracy achieving a 91,07% %; in [23], Runyuan Zhang, observed that using wider, deeper and higher resolution CNN could obtain a better performance, then he proposed a model based on automated melanoma detection by analysis of skin lesion images using EfficientNet B6, which is capable to capture more fine-grained features, obtaining 0.917 AUC-ROC Score. In addition, in [24], authors used convolutional SNNS with an unsupervised spike timing dependent plasticity (STDP) learning rule to classify melanoma and benign skin lesions, and it is proposed to use feature selection to obtain more diagnostic features, improving the networks' classification performance, and achieving an average accuracy of 87.7 %.
The authors of [25], perform melanoma segmentation by using a deep convolution network on hyperspectral pathology images; to take advantage of the properties of three-dimensional hyperspectral data, they proposed a 3D network named Hypernet to segment melanoma from hyperspectral pathology images; to improve the sensitivity of the model, they modified the loss function with caution about the presence of false negatives in diagnosis.
[26] proposed a classification technique based on the ensemble of advantages and a group decision in dermoscopic images, including the ensemble strategy of group decision, the strategy of maximizing individual advantage, and the strategy of block integrated voting. Additionally, using generative adversarial networks (GANS) to build a balanced sample space will produce better training of the convolutional neural networks (CNNS).
Renzo Alberto Villanueva, in [27], proposes an intelligent system based on neural networks to identify melanoma skin cancer in images of skin lesions. This system is composed of 4 modules: preprocessing, where some data augmentation takes place, image classification to analyze images via CNN (EfficientNet B6), contextual information classification to analyze metadata from the images (categories or numbers); and the web interface, obtaining a performance probability of 92.85 % Accuracy, 71.50 % Sensitivity, 94.89 % Specificity.
Due to the interference generated by body hair and its shadows on the skin, the analysis of dermos-copic images presents some challenges to giving a diagnosis. In [28], Lidia Talavera-Martínez shows a new approach to remove those distractors from the image based on machine learning techniques, using an encoder-decoder architecture model with CNN to detect and subsequent restoration of the hair pixels from the images. They also introduce a new loss function that combines L1 distance, total variation loss, and a loss function based on the metric from the index of structural similarity.
In [29], the authors state that lesion change detection is a task that measures the similarity between two dermoscopic images taken over a short period. To achieve this, a deep network based on a Siamese structure is proposed to produce the decision (changed or unchanged). In addition, a tensorial regression process is proposed to extract the global features of the lesion images, together with the deep convolutional features. On the other hand, a segmentation loss (SegLoss) is designed to mimic the decision-making process of doctors. Finally, to evaluate the proposed method, an internal dataset with 1000 pairs of lesion images taken at a melanoma clinical center was established.
In [30], Rozita Rastghalam proposes a detection algorithm based on decision-level fusion and a Hidden Markov Model (HMM), whose parameters are optimized by Expectation Maximization (EM) and asymmetric analysis. The texture heterogeneity of the samples is determined by asymmetric analysis, and in addition, a fusion-based HMM classifier trained with EM is presented, for this, a texture feature is extracted based on local binary patterns, called local difference pattern (LDP) and features of the statistical histogram of the microscopic image.
The framework proposed by Muhammad Attique Khan in [31] consists of two modules, one for lesion segmentation and another for classification. For segmentation, a hybrid strategy is proposed that merges the binary images generated from the designed 16-layer convolutional neural network model with a saliency segmentation based on the high-dimensional contrast transformation (HDCT) enhanced, and a maximum amount of information method is proposed to take advantage of the maximum information extracted from the binary images. In the classification module, a previously trained DenseNet201 model is retrained on the segmented lesion images using transfer learning. Subsequently, the features extracted from the two fully connected layers are down-sampled using the t-distribution stochastic neighbor embedding method and finally, the resulting features are merged using a multiple canonical correlation approach and passed to a multi-class ELM classifier.
Materials and methods
Dataset
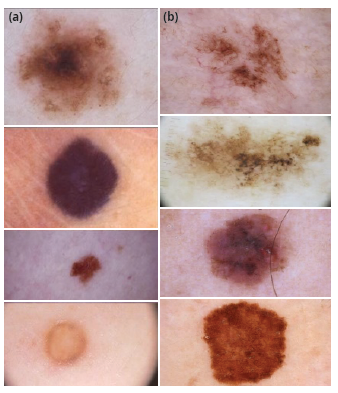
The set of images used to develop this paper was taken from the database fed by the International Skin Imaging Collaboration Archive (ISIC Archive), known as an academia and industry partnership designed to simplify the application of digital skin imaging to help reduce melanoma mortality [5]. This archive contains over 60 000 images from different patients with skin lesions (Figure 1) that could or could not be melanoma, each with its own metadata describing some features from the image such as the patient's approximate age, anatomic location, whether it is benign or malignant, and sex, among others. Although all this information is not available for each image, it is great to find that most of the images are classified as benign (more than 47 000) or malignant (more than 5000).
Proposed Architecture
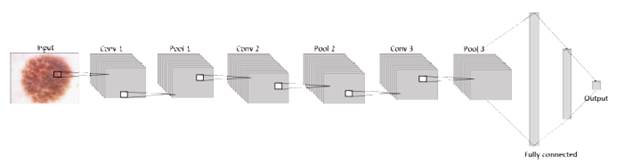
The general architecture handled on each model proposed in this paper is formed by an input layer that contains the image, a system that will be useful for researchers in future projects in areas such as machine learning or others related. It means that at the first layer, the neurons are receiving the information from the images in three dimensions: width, height, and depth (RGB) [32]. Next to the input, the network consists of an exact number of hidden layers, all convolutional layers with pooling and dropout stacked together to extract as many patterns as possible from the image and learn them, before finally using this information in the fully connected layer where the classification is given according to the activation function sigmoid in terms of 1 or 0, as shown in figure 2.
The network counts a set of images, all mixed benign and malignant, taken from the proposed database's 53 310 photos, a portion of which will be randomly selected to build the training, validation, and test datasets, ensuring that no duplicates exist. Sets must be preprocessed to ensure all images have the same height and width. This way, the CNN gets as input a set of images whose dimensions will be understood as height, width, and depth, where the last one belongs to the image's RGB components.
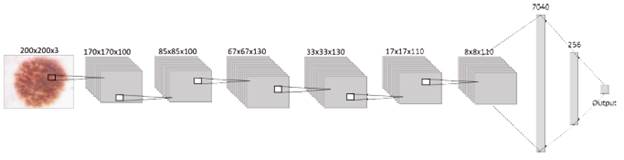
Following the structure shown in Figure 3, next to the input, the proposed CNN structure contains the hidden layers, which are three con-volutional and pooling layers, five dropouts, one flat layer, and two dense layers stacked together. This way, the network has a total of 16 layers that were trained with 33 001 images, where 30 001 are benign injuries and 3000 are melanomas. Layers are in charge of extracting and learning patterns from the input, mainly with convolution, a mathematical linear operation between matrices, the dot product between the weights, and a small region in the input. Then the results are applied to a nonlinear activation function [33], [34], in this case, the ReLU function is applied to every convolutional layer.
After a convolutional layer usually comes a pooling layer, mainly used to reduce dimensions from the previous layer. That is the reason why these two layers (convolutional and pooling) are commonly stacked together, chasing the maximum value (max pooling) in the subregion to get most relevant information to pass it to the next layer [34]. In this paper, 2x2 is the selected size for the pooling layers.
Finally, the classifier in CNN is composed of a fully connected net built with one or more layers, where all neurons in the previous layer are connected to every other neuron in the current layer, which is followed by an output layer [33], where the classification is finally given by the activation function. This paper proposes to use sigmoid to get the output 1 or 0 to classify the lesion as malignant or benign.
Considering how the general behavior of every component of a CNN into account, will make it easier to understand the performance of the structure designed for this paper Beginning with the input layer, which contains data from the images that will be processed by the network. Then, this information is sent through the 3 convolutional layers with the activation function "ReLU," each followed by a pooling layer and dropout, to get this way the most relevant information from the corresponding batch of images and finally flatten it into a 1D vector, which is read by a dense or output layer, where the activation function "Sigmoid" is in charge of realizing the classification, building this way the proposed convolutional neural network as shown in figure 3.
Experimental setup
The data set used to feed the designed network was found on the International Skin Imaging Collaboration (ISIC) web page [5] Firstly, it was a set of more than 23 000 images used in the first phase, but currently it has 53 310 dermoscopic images of different skin lesions, some of which are benign (more than 47 000 images) and others are melanomas (more than 5000). Considering the limited set of images, it was necessary to use most of the set to train the network, distributed as follows: a training set of 33 001 images (approx. 61 % of the dataset) and 11 500 for the validation set (approx. 21 %), keeping some others to test the CNN behavior. Images must be passed through preprocessing to resize them to 200x200, making sure they all have the same size at the input of the network.
The machine used to train the network has the following characteristics: Linux Mint as an operative system, Intel Core i7 processor, 0GB RAM, a 1TB HDD, and an NVIDIA GeForce GTX 1050 with 4GB of GDDR5 dedicated VRAM. Also, the proposed architecture was developed by using Keras and TensorFlow, both Python open-source libraries dedicated to machine learning processes.
Experiment and results
Following the experimental setup, to understand the results of this paper, it's necessary to understand how both phases worked. During the development of the first phase, the data set used was small, with around 23 000 images, and, considering that the number of images per class was so marked (around 10 benign lesions per one melanoma), it turned the experiments into a game around the quantity of images to train, validate, and test. This way, the first experiment only had 60 images to test, the second one was trained with around 3000 images of benign lesions and melanomas distributed 50/50, and based on those two experiments, for the rest of the cases in that phase, they were trained t o keep the distribution of the set of images at 70 for training and 30 for validation, and for testing it was necessary to do it with the same validation set. Then, results obtained in this phase are not 100 % trustworthy due to the way the models were tested. Knowing that most of the results in this phase gave accuracy percentages around 90 %, but those numbers were because most of the images in the set test are not melanomas, it means that even if the model fails all the melanoma cases, it will always get an accuracy percentage close to 100.
Based on the experiments realized in phase one and the unreliable results obtained, to solve the problem due to the notorious disparity in the quantity of the images in the data set, in this case, it was proposed to duplicate the images of melanoma cases as many times as it was necessary by copying and pasting the set of images of malignant lesions. Several times until getting the same quantity of images as for benign lesions, this models will handle balanced classes for training and validation.
The experiments, like phase one, consist of a CNN trained with preprocessed images; this step ensures that all images are the same size so that the neural network can be trained by extracting patterns with three convolutional layers, pooling, and dropout stacked together, but with the difference that for those experiments, the training, validating, and testing sets will remain immutable. This way, the second phase experiments were focused on parameters like the learning rate, dropout, depth, kernels, and pooling size, as shown in Table 3, while keeping others fixed, like in Table 2 and always avoiding overfitting. To get the classification as zero or one, the activation functions used were ReLU for each layer and Sigmoid at the output layer of the fully connected layer.
Table 2 Fixed parameters used to train the architecture and test results.
| Epochs | 10 |
| Image size | 100x100 |
| Batch size | 100 |
| Training steps | 600 |
| Validation steps | 190 |
| Number of convolutional layers | 3 |
| Learning rate | 1x10^-5 |
| Activation function | ReLU |
| Sigmoid in the output layer |
Source: The authors.
In Table 3, variable parameters are shown as they were applied in the first 4 experiments of the second phase to train the models, but for the fifth experiment, the best three models were selected from the last experiments based on the accuracy obtained on tenting, and, those cases were trained to change the pooling size for each convolution layer starting with 3x3.
Table 3 Configurations from the first 4 experiments.
| Experiment | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Conv1 | 100-7-7 | 70-7-7 | 200-7-7 | 200-9-9 |
| Conv2 | 130-7-7 | 140-7-7 | 150-7-7 | 150-9-9 |
| Conv3 | 110-3-3 | 100-5-5 | 100-3-3 | 100-7-7 |
| Dropout | 0.2- 0.3- 0.4 | 0.2- 0.3- 0.4 | 0.2- 0.3- 0.4 | 0.2- 0.3- 0.4 |
Source: The authors.
The proposed architecture was trained on each model per 10 epochs with the different combinations from Table 3 to find the model with the best results and, evaluate the performance of all the models previously described in Table 2 and Table 3. Using a test set with 8,809 images, both melanoma and nonmelanoma, the highest accuracy found was 88.75 %. However, from the 15 models trained, three were selected due to their results evaluated through a confusion matrix and f1 score, as shown in Table 4.
Table 4 shows the results obtained in models 5, 7, and 12, all of them with accuracy percentages greater than 84 %, which is superior to the lowest accuracy reached for a dermatologist with experience. Based on the confusion matrices, analyzing them one by one, model 5 has the highest accuracy but also the lowest specificity, which means the model will give more false benign cases than other models, which means it will have a high number of melanomas misclassified, which is not desired because many cases of this skin cancer will not get treatment in the early stages.
Opposite to the last model, in model 7, the specificity is the highest, reducing the number of melanomas misclassified and, on the way, sacrificing accuracy and sensitivity. In that situation, more nonmelanoma lesions will be misclassified as malignant melanoma, and then more unnecessary biopsies will be made, going against of one the purposes of this tool. In model 12, it was possible to get a middle point with specificity greater than 60 %, sensitivity greater than 90 %, and accuracy of 87.82 %, mispredictions in both scenarios in that case. Beyond having more stable results than in phase one, other metrics support the results in the last model, like the PPV (Positive Predictive Value) or NPV (Negative Predictive Value), which confirm that model 94.43 % of the benign predictions are truly benign lesions and 51.93 % of the malignant predictions are effectively melanomas, which means this model not only obtained a high number of well-classified melanomas, but also a reduced number of mis-classified benign lesions compared with the other models and at the same time reduces the number of unnecessary biopsies.
On the other hand, the likelihood ratio values, in this case, are lk+ = 2487 and lk- = 0.136, which means that in an evaluation posttest, a prediction is 2487 times more likely that the classified lesion as nonmelanoma is currently a benign lesion, while with lk-, the probability to obtain a benign prediction from a malignant case decreases. This method provides additional tools to medical professionals in order to provide a more reliable screening. Finally, the f1-score supports the choice of the model due to the fact that it evaluates the performance of precision and recall, combined giving both metrics the same importance in this case with a value of 0.929.
Table 4 Confusion Matrix Model 706, evaluating with the test set.
Source: The authors.
Conclusion and future works
In this paper, a non-trivial architecture of a CNN is proposed to get a trustworthy diagnosis for malignant melanomas by applying the ReLU function in all convolutional layers and the sigmoid function at the output layer to finally obtain the classification. However, due to the limited data set, a great number of similarities between melanomas and benign lesions, and the apparent difference in the number of malignant lesions images compared to the benign ones, it was mandatory to duplicate several times the set of malignant photos to obtain this way more stable and trustable results comparing both phases. The system then developed is capable of detecting cases of melanomas among non-melanomas cases with an accuracy of 87.82 %. It was also necessary to use another kind of metric to evaluate the trained CNN and get a more confident result. Due to this is possible to notice that even when the show results are not the highest, they are steady and provide a low quantity of false positives and false negatives, demonstrating higher accuracy in their performance than the human eye.
Another of the encountered limitations is the hardware employed to train the models designed, due to the lesions images that maybe are not all clear enough, the model complexity that demands a better processor, lots of hours invested on pieces of training because every training with a reasonably deep network took around 2 days to process ten epochs and its respective validation, and the machine's capacity itself, where the training process of a deeper network, by needing a greater machine's capacity, crashes and stops the process, never converging the learning curve.
Compared with other systems developed by the community, shown in section previous works and Table 5, it is possible to notice that the results obtained are not the best. This due to some main differences found, such as machine specifications, where some networks were trained with more powerful GPUS than the GTX 1050 or even those machines count with more than one GPU; and on the other hand, those projects also applied some data argumentation tools at the preprocessing stage and other classification methods or loss functions like Support Machine Vector or Fisher Vector. However, it is noticed that even with limited machine capacities, the proposed architecture and the image distribution used made it possible to get satisfactory results, which probably could be better if a better machine and other techniques were implemented, such as those exposed in other works mentioned in the previous works section.
Table 5 Results of other systems for melanoma detection
| Dataset | Ref | Acc | AUC | F1 | Machine Specifications |
|---|---|---|---|---|---|
| ISBI 2016 | [14] | 95.5 % | - | - | Dell XPS 8900 desktop with Intel(R) i7 6700 3.4 GHz CPU GPU of Nvidia GeForce GTX 1060 with 6GB GDDR5 memory |
| ISBI 2016 | [13] | 85.5 % | 0.78 | - | NVIDIA TITAN X GPU |
| CNN's | [16] | 81.00 % | - | - | Intel Core i7-4790K 32 GB - NVIDIA GeForce GTX Titan X GPU cards with scalable link interface (SLI) |
| PH2 | [35] | 96.7 % | 0.9949 | 0.9948 | 2.8 GHz 16 BG GPU support of 4 GB |
| ISBI 2016 | [36] | 92.1 % | 0.97 | - | The proposed model is implemented in on MATLAB 2017b to perform experiments using high-processing GPU systems |
| ISBI 2016 | [37] | 86.81 % | 0.852 | - | Inter Xeon E5 2680 @ 2.70 GHz 128GB NVIDIA Quadro K4000 |
| ISBI 2017 | [38] | 85.3 % | - | - | NVIDIA TITAN X GPU |
| ISBI 2017 | [39] | 95.86 % | - | - | Intel(R) Core(TM) i7-7700HQ CPU 2.80 GHz Nvidia GTX 1050 Ti GPU |
| ISBI 2017 | [36] | 96.5 % | 0.99 | - | The proposed model is implemented in MATLAB 2017b to perform experiments using high-processing GPU systems |
| ISBI 2017 | [40] | 83.00 % | 0.842 | - | NVIDIA GTX1080 GPU with 8GB of memory |
| ISBI 2017 | [41] | 85.7 % | 0.912 | - | GeForce GTX TITAN X, 12 GB RAM |
| ISBI 2018 | [36] | 85.1 % | 0.92 | - | The proposed model is implemented in MATLAB 2017b to perform experiments using high-processing GPU systems |
| ISIC Archive | [35] | 99.5 % | 0.994 | - | GPU support of 4 GB |
| Med-Node | [35] | 95.2 % | 0.944 | - | GPU support of 4 GB |
| ISCI2016 | [42] | 86.3 % | 0.861 | 0,893 | Intel(R) Core (TM) i7-7820X CPU @ 3.60GHz RTX 2080Ti GPU |
| ISIC 2017 | [42] | 85.00 % | 0.891 | Intel(R) Core (TM) i7-7820X CPU @ 3.60GHz RTX 2080Ti GPU | |
| DermIS | [43] | 97.9 % AUG - 81.30 % | - | 0,9770 | 6GB NVIDIA GPU card |
| DermQUEST | [43] | 97.4 % AUG - 74.2 % | - | 0,9790 | 6GB NVIDIA GPU card |
| IS-BI2017 | [44] | 95.70 % | - | - | Intel i7 7700k CPU 32 GB DDR4 RAM Nvidia GeForce GTX 1080Ti (11 GB) |
| ISIC2018 | [44] | 94.70 % | - | - | Intel i7-7700k CPU 32 GB DDR4 RAM Nvidia GeForce GTX 1080Ti (11 GB) |
| ISIC 2017 | [45] | 94.96 % | - | - | Intel Xeon (R) CPU E5 2620 v3 2.40GHz NVIDIA Tesla K80 (12G)GPU |
| PH2 | [45] | 95.23 % | - | - | Intel Xeon (R) CPU E5 2620 v3 2.40GHz NVIDIA Tesla K80 (12G)GPU |
| ISIC 2018 | [46] | 92.60 % | 0.918 | 0.491 | Four NVIDIA Tesla P100 GPUs with 16GB of memory |
| ISIC 2019 | [46] | 92.40 % | 0.919 | 0.488 | Four NVIDIA Tesla P100 GPUs with 16GB of memory |
| ISBI 2016 | [47] | 87.60 % | 0.854 | - | NVIDIA Tesla K80 GPU (12G) |
| ISIC 2017 | [48] | 85.00 % | 0.875 | - | ARLCNN50 model with one NVIDIA GTX Titan XP GPU |
Source: The authors.
On the other hand, the visual evaluation made by a specialist in dermatology is considered a medical screening based on the inconveniences presented in past experiments. Furthermore, rather than using the proposed architecture and method of duplicating images to obtain an even distribution between the classes in future applications, it is planned to use a different technique to provide more clear images, previously processed of the skin lesion (benign or malignant), and other data related to the image (metadata), pointing to get higher accuracy and lower loss percentages, as well as higher F1 Score, giving the specialist a prediction as close as possible to the real diagnosis, based on a system trained from a convolutional neural network using more specific images without unnecessary data and information previously collected and analyzed by others to understand the development of that kind of cancer in human beings, like probabilities for a man or a woman at certain ages to suffer it, knowing that, by definition, it is not possible to get an exact diagnosis of melanoma until the biopsy.