Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Facultad Nacional de Agronomía Medellín
Print version ISSN 0304-2847
Rev. Fac. Nac. Agron. Medellín vol.62 no.2 Medellín July/Dec. 2009
CHARACTERIZATION OF Phytophthora infestans POPULATIONS IN ANTIOQUIA, COLOMBIA
CARACTERIZACIÓN DE LAS POBLACIONES DE Phytophthora infestan EN ANTIOQUIA, COLOMBIA
Elizabeth Gilchrist Ramelli1; Sonia Jaramillo Villegas2; Lucia Afanador Kafuri3 and Rafael Eduardo Arango Isaza4
1 Ingeniera Agrónoma, D.Sc. Universidad Nacional de Colombia, Sede Medellín. Facultad de Ciencias Agropecuarias. A.A. 1779. Medellín, Colombia. <elygilchrist@hotmail.com>
2 Profesora Asociada. Universidad Nacional de Colombia, Sede Medellín. Facultad de Ciencias Agropecuarias. A.A. 1779. Medellín, Colombia. <sjaramal@unalmed.edu.co>
3 Profesora Asociada. Universidad Nacional de Colombia, Sede Medellín. Facultad de Ciencias Agropecuarias. A.A. 1779. Medellín, Colombia. <lafanado@unal.edu.co>
4 Profesor Asociado. Universidad Nacional de Colombia, Sede Medellín. Facultad de Ciencias Agropecuarias. A.A. 1779. Medellín, Colombia
Recibido: Abril 4 de 2008; Aceptado: Septiembre 7 de 2009.
Abstract. From the Phytophthora infestans collection of the Universidad Nacional de Colombia, the isolates collected in different locations in Antioquia, Colombia between 1994 and 2000 were evaluated. These isolates were obtained from late blight lessons in different hosts. In 2000, these isolates were characterized by mating type, mitochondrial haplotype and virulence races. All isolates were of the A1 mating type and two mitochondrial haplotypes were identified: IIa, present in isolates from all the hosts tested, and Ib present only in isolates from tomato and water cucumber (Solanum muricatum). The Antioquia population of P. infestans showed a large complexity of virulence factors (10 out 11), especially those isolates collected from potato, while the tomato population was less complex. The A1 mating type and the mitochondrial haplotype IIa has been associated with the EC1 population that possibly is replacing the US1 population.
Key words: Late blight, clonal lineage, South America, haplotypes, mating type.
Resumen. De la colección de Phytophthora infestans de la Universidad Nacional de Colombia, se evaluaron aquellos aislamientos provenientes de diferentes localidades de Antioquia, Colombia entre 1994 y 2000. Dichos aislamientos fueron obtenidos de lesiones de tizón tardío en diferentes hospederos. En el año 2000 se caracterizaron por el tipo de apareamiento, haplotipo mitocondrial y razas de virulencia. Todos los aislamientos correspondieron al tipo de apareamiento A1 y se presentaron dos haplotipos mitocondriales: IIa, en aislamientos de todos los hospederos evaluados, y Ib solamente en aislamientos colectados de tomate y pepino de agua (Solanum muricatum). La población antioqueña de P. infestans presenta una amplia complejidad de factores de virulencia (10 de 11), especialmente para los aislamientos colectados de papa, mientras que la población de tomate fue menos compleja. El tipo de apareamiento A1 y el haplotipo mitocondrial IIa han sido asociados a la población EC1 que posiblemente está desplazando la población US1.
Palabras claves: Tizón tardío, linaje clonal, Sur América, haplotipos, grupo de compatibilidad.
The oomycete Phytophthora infestans (Mont.) de Bary is the causal agent of several economically important diseases in potato and tomato. In tropical countries the pathogen infects other crops like water cucumber (Solanum muricatum), tamarillo (S. betaceum) and lulo (Solanum quitoense). The population of P. infestans is mostly composed of clonal lineages with distinct genotypes that can be characterized by RLFP’s (Restriction Fragment Length Polymorphism), isozimes and mitochondrial haplotypes (Oyarzun et al., 1997; Pérez et al., 2001).
P. infestans reproduces sexually if the two mating types A1 and A2 are present. Sexual reproduction is present in the entire world. It has been reported in Mexico (Forbes et al., 1998; Garry et al., 2005), Europe (Drenth et al., 1994) and Asia (Ghimire et al., 2003). It is also present in South América, specially in Venezuela (Briceño et al., 2009) and in Colombia (Vargas et al., 2009). The evidence confirms the asexual reproduction of P. infestans, since only one isolate of 840 collected in Venezuela presented fertility with A1. In Colombia, Vargas et al. (2009) found the A2 mating type in one isolate collected from Physalis peruviana L. However, this isolate generates oospores with the A1 mating type reference isolate, but this oospores are not infective (Restrepo Silvia, Universidad de los Andes, personal communication). In Ecuador, the US-1 genotype has been replaced by the EC-1 lineage, which is more pathogenic and less sensitive to the systemic fungicide metalaxyl (Pérez et al., 2001; Forbes et al., 1996). In Colombia, US-1 and EC-1 clonal lineages have been reported (Griffith and Shaw, 1998; Forbes et al., 1998; Vargas et al., 2009).
The populations of P. infestans can be also studied in terms of their physiological races which are determined by the presence of one or more of the 11 known virulence genes. No correlation between physiological race and genotype has been found so far and several studies indicate that the complexity of virulence factors in the oomycete has increased with time (Tooley et al., 1986). At the time of this research (1994-2000), there was a small amount of information available concerning the Colombian populations of P. infestans and the strategic geographical position of this country justifies a study on the structure of the population of this pathogen. Forty isolates of P. infestans collected from different hosts and locations in the north-central Colombian Andes (Antioquia department) were characterized by mating type, mitochondrial haplotype and physiological races. This work, carried out in 2000, enriches the study of populations of the pathogen in the South American continent and the rest of the world.
MATERIALS AND METHODS
Sources of Phytophthora infestans isolates. Between 1994 and 2000, 40 isolates of P. infestans were collected from affected plants of commercial growing fields and house garden of potato Solanum tuberosum cultivars (ICA Puracé, ICA Nevada, ICA Cumanday and Diacol Capiro), Solanum phureja, tomato and water cucumber. Isolates were taken from infected leaf tissue and collected in 14 different locations of the Antioquia department. Geographical origin (First letter of each isolate, A: Antioquia), year (last two numbers of each isolate), and host of each isolate were recorded (Table 1; Figure 1).
Table 1. Localization of Phytophthora infestans isolates from Antioquia-Colombia used in this study. Location from wich isolates were collected. Mating type, virulence factors, mitochondrial haplotype and clonal lineage for each isolate are indicated.
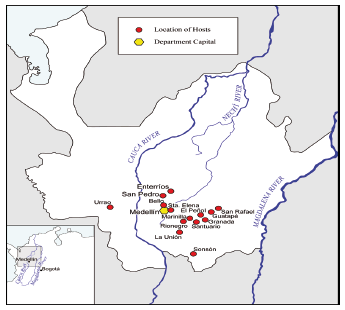
Figure 1. Map of Antioquia-Colombia, showing detailed locations where isolates of Phytophthora infestans were collected.
Media and culture conditions. All isolates of P. infestans were kept at 15-18 ºC on Petri dishes containing rye agar (25 g of rye powder, 20 g of table sugar and 18 g of agar per liter). For the sexual compatibility assay the media was complemented with 0.05 g/l of b-sitosterol. For DNA extraction, mycelia were grown for 8 days in pea water broth (30 g of frozen peas and 20 g of table sugar per liter), harvested by filtration and stored at –20 ºC.
Mating type. To avoid the introduction to the country of unknown mating type, at the date of this research, only the mating type A1 was used as a control. Mating type was determined by paring each isolate with a known A1 culture on rye-agar medium (Deahl et al., 1991). Plates were incubated at 15 ºC in the dark, during 30 days. Analysis consisted on checking each pairing for the formation of oospores under macro and microscopic examinations of micelia formed in the area of interaction between the isolates paired. A self-paring of each isolate was used as a control.
DNA extraction. DNA was extracted using the method of Afanador et al. (1993). Briefly, 100 mL of mycelium powder was incubated in a microcentrifuge tube with 400 mL of CTAB extraction buffer (CTAB 2% w/v, 100 mM Tris HCl, pH 8.0, 1.4 M NaCl, 20 mM EDTA pH 8.0), for 30 min at 65 ºC, inverting the tubes every 8 min. Additional 400 mL of cloroform:isoamyl alcohol (24:1) were added, mixed by inversion during 15 min, followed by centrifugation at 20.000 CRF (Centrifugal Relative Force) during 10 min. This step was repeated, and followed by precipitation with isopropanol and resuspention in TE buffer.
Mitochondrial haplotypes. Mitochondrial haplotypes were determined by PCR-RFLP using primer pairs P1 (P1 forward and P1 reverse), P2 (P2 forward and P2 reverse) and P4 (P4 forward and P4 reverse) as described by Griffith and Shaw (1998). PCR (PTC-100TM MJ Research, Inc.) amplification conditions were as follows for all primer combinations: deoxynucleotide triphosphates dNTP´s (Promega), 200 mM each, PCR buffer 1X (1 M KCl, 1 M Tris HCl pH 8.3, 1 M MgCl2), primers (Operon Technologies, Inc.) 0.34 mM each, bovine serum albumin, 160 mg/mL, Taq DNA polymerase (Promega) 0.2 mL (1U) and 2 a 10 ng of total DNA in 0.2 mL microcentrifuge tubes (final volume, 25 mL). PCR cycles were as follows: 1 cycle of 94 ºC for 90 s; 40 cycles of 94 ºC for 40s, 55 ºC for 60s and 72 ºC for 120s.
Five microliters of the amplified product was digested with the following restriction enzymes (Promega): P1 reactions with CfoI, P2 with MspI and P4 with EcoRI. Digested DNA (10 mL) was mixed with 5 mL of loading buffer and loaded into a 2% agarose gel in 1 X TBE buffer (containing 0.1 mg/mL of ethidium bromide). The gel was run at 80 vol. during 4 h. Restriction patterns were visualized with an UV transilluminator and the images were recorded by a gel documentation system (BIO-RAD, Gel Doc 1000).
Physiological races. The virulence specificity of a subset of 28 isolates was determined by inoculation of detached leaflets from the international set of differential potato cultivars provided by the Centro Internacional de la Papa (CIP), carrying the 11 known mayor (R) resistance genes. Briefly, leaflets collected from the middle part of each cultivar were inoculated in a moist chamber with 20 mL of sporangial suspension (approx. 5.000 sporangia/mL) placed on the abaxial surface of each leaflet. Each test included the susceptible cultivar Alfa (a) (containing no know R genes) as a control. The virulence assays were repeated at least twice.
RESULTS
Mating type. In all tests only mating type A1 was detected as indicated by the absence of oospores in the growing area between the isolates on all pairings.
Mitochondrial haplotypes. PCR-RFLP analysis revealed the presence of two mitochondrial haplotypes in the Antioquian population of P. infestans: haplotypes IIa and Ib (Table 1). The mitochondrial haplotype IIa was present in all potato isolates (29 samples), in 2 isolates from tomato and in 3 isolates from water cucumber, while the haplotype Ib was only present in 5 isolates from tomato and in one isolate from water cucumber (Figure 2).
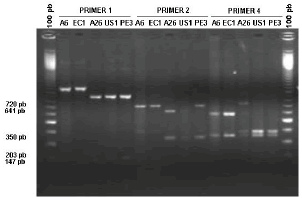
Figure 2. Mitochondrial haplotypes of Phytophthora infestans, as analyzed by primer pairs P1, P2 y P4. From left to right 100 pb weight marker, A6, EC1, A26, US1, Pe3 samples amplified with P1, P2 and P4 primers, 100 pb weight marker.
Physiological races. Eighteen pathotypes were identified among 28 isolates tested, showing variation in the level of complexity of their races. The most common pathotype was presented on 21.42 % of the isolates, and infected ten of the eleven potato cultivars (1.2.3.4.6.7.8.9.10.11). One of the isolates coming from tomato had no virulence genes (Race 0). Virulence factor No. 5 was not present in any isolate. Furthermore, the isolates collected after 1997, infected the R9 cultivar, which indicates an increment in the pathogen complexity. The R3 cultivar was the most affected by the tested isolates, by a 26/28 proportion. To calculate the Shannon (HS) and Gleason (HG) diversity index, isolates were divided in two groups by date of collection: one was from 1994 to 1996, and the other from 1997 to 2000. This analysis showed an increase on diversity factors from HG =2.5 and HS=1.47 to HG =4.26 and HS=2.09.
DISCUSSION
Forty isolates of P. infestans were analyzed in this work, which were collected from different Solanum hosts and locations of a north-central Colombian Andean region of the Antioquia department, South America. The isolates were tested for mating type, mitochondrial haplotype and virulence factors.
All isolates tested showed to be of the mating type A1 suggesting that the current population of P. infestans in Antioquia is clonal with asexual reproduction. To 2000, the presence of the A2 mating type of P. infestans in Colombia cannot be ruled out considering that is has been reported in Ecuador, in association with wild species of Solanum such as S. brevifolium y S. tetrapetalum (Ordoñez et al., 2000), in potato in Uruguay, Brazil, Bolivia and Argentina (Brommonschenkel 1988; Deahl et al., 2003), and in Mexico and the United States (Deahl et al., 1991; Goodwin et al., 1992). In recent studies the A2 mating type has been reported in Colombia in P. peruviana (Vargas et al., 2009).
These results suggest that it might be important to use a different sampling strategy to continue monitoring the Colombian population of P. infestans, increasing the range of hosts for sampling. This approach, combined with regulations on the importation of asexual seeds and the control of other host species, may avoid or retard the entrance of the infective A2 mating type and the probability to produce infective oospores with the A2 isolates currently present.
Two mitochondrial haplotypes of P. infestans are present in the population studied, the IIa haplotype which is related with the clonal lineage EC-1 (Oyarzun et al., 1997), and the Ib related with the US-1 (Griffith and Shaw, 1998). This finding is consistent with what has been found in Peru, where EC-1 populations have been replaced by US-1. This change in the population is probably due to the fact that EC-1 is resistant to the systemic fungicide metalaxyl, whereas US-1 is sensitive to it (Pérez et al., 1998). In Marruecos, Sedegui et al. (2000) found changes on P. infestans population in potato, but not in tomato, which is in agreement with the results obtained in this study.
As seen in Table 1 the complexity of the P. infestans population in potato is higher that the one in tomato, and this could be associated with clonal lineage US-1 being less complex than EC-1 as has been found in Ecuador (Forbes et al., 1997) and in Peru (Pérez et al., 2001). Several works worldwide have shown that the isolates that correspond to the so called “new” (EC-1) population are more aggressive on their original host (Goodwin et al., 1995; Lebreton et al., 1999; Sedegui et al., 2000), indicating that the pathogen is coevolving with its host and adapting to the agricultural practices that farmers are using in each region, such as the type of chemical control employed and the genetic make up of the cultivars. Furthermore, extensive potato crops with a single variety or a very narrow genetic base are highly susceptible to any adaptation in the microorganism.
From this study we can conclude that the P. infestans population in Antioquia-Colombia up to 2000, was A1 mating type, composed mainly of the two mitochondrial haplotypes, IIa and Ib which are associated with the clonal lineages EC-1 and US-1, respectively. The clonal lineages present were very diverse on its virulence factors, specially the EC-1 population, which is more complex than the US-1 lineage. Our work contributes to the worldwide effort to characterize P. infestans and it specially gives some information on the pathogen populations in a place located in a strategic region in the South American continent, since Colombia is the crosspoint between South and Central America.
ACKNOWLEDMENTS
This research was supported in part by COLCIENCIAS, Universidad Nacional de Colombia, Sede Medellín and FEDEPAPA. Special thanks to José Manual Gonzalez, for their cooperation with the map design.
BIBLIOGRAPHY
Afanador, L., S.D. Haley and J.D. Kelly. 1993. Adoption of a “mini-prep” DNA extraction method for RAPD marker analysis in common bean (Phaseolus vulgaris L.). Annual Report of the Bean Improvement Cooperative 36: 10-11.
Black, W, C. Mastenbroek, W.R. Mills and L.C. Peterson. 1953. A proposal for an international nomenclature of races of Phytophthora infestans and genes controlling immunity in Solanum demissum derivates. Euphytica 2(3): 173-179. [ Links ]
Briceño, A, M. San Román, M. Moreno, G. Fermin, L. Cedeño, K. Quintero and H. Pino. 2009. Population structure of Phytophthora infestans in the Venezuelan Andes (2004-2007). Acta Horticulturae (ISHS) 834:129-140. [ Links ]
Brommonschenkel, S.H. 1988. Patogenicidae, compatibilidae, citogenética e padroes isoenzimaticos de isolados de Phytophthora infestans (Mont) De Bary do Brasil. Tese de Mestrado. Universidade Federal de Vicosa. Minas Gerais. Brasil - Janeiro. 82 p. [ Links ]
Deahl, K.L., R.W. Goth, R. Young, S.L. Sinden and M.E. Gallegly. 1991. Occurrence of the A2 mating type of Phytophthora infestans in potato fields in the United States and Canada. American Journal of Potato Research 68(11): 717-725. [ Links ]
Deahl, K.L., M.C. Pagani, F.L. Vilaro, F.M. Perez, B. Moravec and L.R. Cooke. 2003. Characteristics of Phytophthora infestans isolates from Uruguay. European Journal of Plant Pathology 109(3): 277–281.
Drenth, A., I.C.Q. Tas and F. Govers. 1994. DNA fingerprinting uncovers a new sexually reproducing population of Phytophthora infestans in the Netherlands. European Journal of Plant Pathology 100: 97–107.
Forbes, G.A., P.J. Oyarzún, A. Poso and M.E. Ordóñez. 1996. Host specificity of late blight pathogen on potato and tomato in Ecuador. En: Centro Internacional de la Papa, http://www.cipotato.org/market-PgmRports/pr95-96/program3-prog35.htm; consulta: abril 2008. [ Links ]
Forbes, G.A., X. Escobar, C. Ayala, J. Revelo, M.E. Ordoñez, B.A. Fry, K. Doucett and W.E. Fry. 1997. Population genetic structure of Phytophthora infestans in Ecuador. Phytopathology 87(4): 375-380. [ Links ]
Forbes, G.A., S.B. Goodwin, A. Drenth, P. Oyarzun, M.E. Ordoñez and W.E. Fry. 1998. A global marker database for Phytophthora infestans. Plant Disease 82(7): 811-818. [ Links ]
Garry, G., G.A. Forbes, A. Salas, M. Santa Cruz, W.G. Perez and R.J. Nelson. 2005. Genetic diversity and host differentiation among isolates of Phytophthora infestans from cultivated potato and wild solanaceous hosts in Peru. Plant Pathology 54(6): 740–748.
Ghimire, S.R., K.D. Hyde, I.J. Hodgkiss, D.S. Shaw and E.C.Y. Liew. 2003. Variations in the Phytophthora infestans population in Nepal as revealed by nuclear and mitochondrial DNA polymorphisms. Phytopathology 93(2): 236–243.
Goodwin, S.B., L.J. Spielman, J.M. Matuszak, S.N. Bergeron and W.E. Fry. 1992. Clonal diversity and genetic differentiation of Phytophthora infestans populations in northern and central Mexico. Phytopathology 82(9): 955-961. [ Links ]
Goodwin, S.B., L.S. Sujkowski and W.G. Fry. 1995. Rapid evolution of pathogenicity within clonal lineages of the potato late blight disease fungus. Phytopathology 85(6): 669-676. [ Links ]
Griffith, G.W. and D.S. Shaw. 1998. Polymorphisms in Phytophthora infestans: Four mitochondrial haplotypes are detected after PCR amplification of DNA from pure cultures or from host lesions. Applied and Environmental Microbiology 64(10): 4007-4014. [ Links ]
Lebreton, L., J.M. Lucas and D. Andrivon. 1999. Aggressiveness and competitive fitness of Phytophthora infestans isolates collected from potato and tomato in France. Phytopathology 89(8): 679-686. [ Links ]
Ordoñez, M.E., H.R. Hohl, A. Velasco, M.P. Ramon, P. Oyarazun, C.D. Smart, W.E. Fry, G.A. Forbes and L.J. Erselius. 2000. A novel population of Phytophthora, similar to Phytophthora infestans, attacks wild Solanum species in Ecuador. Phytopathology 90(2):197-202. [ Links ]
Oyarzun, P.J., A. Pozo, M.E. Ordoñez and G.A. Forbes. 1997. Host specialization of Phytophthora infestans in Ecuador. International Potato Center, Quito, Ecuador. Phytopathology 87:S73. [ Links ]
Pérez, W., S. Gamboa, M. Coca, R. Raymundo, R. Hijmans and R. Nelson. 1998. Characterization of Phytophthora infestans populations in Peru. In: International Potato Center (ed.). Impact on a Changing World. CIP Program Report 1997-1998. [ Links ]
Pérez, W.G., S. Gamboa, Y.V. Falcon, M. Coca, R.M. Raymundo and J.R. Nelson. 2001. Genetic structure of Peruvian populations of Phytophthora infestans. Phythopathology 91(10): 956-965. [ Links ]
Sedegui, M., R.B. Carrol, A.L. Morehart, T.A. Evans, S.H. Kim, R. Lakhdar and A. Arifi. 2000. Genetic structure of the Phytophthora infestans population in Morocco. Plant Disease 84(2): 173-176. [ Links ]
Tooley, P.W., J.A. Sweigard and W.E. Fry. 1986. Fitness and virulence of Phytophthora infestans isolates from sexual and asexual populations. Phytopathology 76(11):1209–1212.
Vargas, A., A. González, D. Garnica, R. Sierra, L.M. Rodriguez, A. Pinzón, M.C. Céspedes, A. Rojas, G. Fermín, M. Marín, L.E. Lagos, A. Bernal y S. Restrepo. 2009. Phytophthora infestans: genética, genómica y bioinformática, la unión hace la fuerza. http://www.gobant.gov.co/organismos/agricultura/papa/cadena%20papa/investigaciones/Phytophthora%20infestans.pdf; consulta: junio 2009. [ Links ]














