Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista Facultad Nacional de Agronomía Medellín
versão impressa ISSN 0304-2847
Rev. Fac. Nac. Agron. Medellín vol.68 no.1 Medellín jan./jun. 2015
https://doi.org/10.15446/rfnam.v68n1.47826
Inter and Intra Variation of Potato Yellow Vein Virus in Three Potato Species From Colombia
Variación Inter e Intra de Potato Yellow Vein Virus en Tres Especies de Papa de Colombia
Patricia Andrea Rodríguez1; Liliana Franco Lara2 and Mónica Guzmán Barney3
1 Biologist. Universidad Nacional de Colombia - Sede Bogotá - Instituto de Biotecnología IBUN. Carrera 45, Calle 26, Bogotá, Colombia. <biopatricia@hotmail.com>
2 Associate Professor. Universidad Militar Nueva Granada - Facultad de Ciencias Básicas y Aplicadas - Grupo de Biología Molecular de Virus. km 2 Via Cajicá - Zipaquirá, Cajicá, Colombia. <liliana.franco@unimilitar.edu.co>
3 Associate Professor. Universidad Nacional de Colombia - Sede Bogotá - Plant Virus laboratory Instituto de Biotecnología IBUN. Grupo de Biología Molecular de Virus- UN. Carrera 45, Calle 26, Bogotá, Colombia. <mmguzmanb@unal.edu.co>
Received: February 05, 2014; Accepted: April 01, 2014.
doi: http://dx.doi.org/10.15446/rfnam.v68n1.47826
Abstract. Potato yellow vein virus (PYVV), (family Closteroviridae, genus Crinivirus) is a re-emergent virus in Andean countries. Low inter-isolate variation has been reported for PYVV CP gene, but there are no reports for intra-isolate variation. Inter- and intra-isolate variability in CP from a population of PYVV was studied. Samples of 216 symptomatic potato plants (115 Solanum tuberosum subsp. andigena (STA), 100 Solanum phureja (SPH) and 1 Solanum chaucha (SCH)) were collected in five Colombian departments. Viral isolates were amplified by RT-PCR and the amplicons were analyzed by single-strand conformation polymorphism (SSCP). Six different migration SSCP patterns (A to F) with different complexities were observed among the population. Pattern A was detected in the five departments in 66% of the isolates. Pattern E was found only in the department of Cundinamarca with a frequency of 0.09%. Patterns B, C, D and F were found in similar proportions of from 13% to 5.6% and were present in the five departments. Homology at the nucleotide level of 75% of the sequence of the CP gene was greater than 99% and the dN/dS ratio (no-synonymous/synonymous changes) was 0.002. Amplicons of the whole CP gene of eight selected isolates representing the six SSCP patterns were cloned and the SSCP analysis showed that, in all cases, more than one variant was present. The sequence analysis of the 35 clones confirmed intra-isolate variability of PYVV. The existence of several variants in a single field isolate was demonstrated and negative selection against amino acid changes of the CP was suggested.
Key words: PYVV, coat protein, intra variability, SSCP.
Resumen. Potato yellow vein virus (PYVV), (familia Closteroviridae, género Crinivirus) es un virus re-emergente para los países Andinos. Se ha reportado baja variación del gen CP de PYVV entre aislados, pero no hay reportes de variación intra aislado. Se estudió tanto la variación inter como intra aislado del CP de una población de PYVV. Muestras de 216 plantas de papa sintomáticas (115 Solanum tuberosum subsp. andigena (STA), 100 Solanum phureja (SPH) y 1 Solanum chaucha (SCH)) se colectaron en 5 departamentos de Colombia. Los aislados se amplificaron por RT-PCR y los amplicones se analizaron por polimorfismo conformacional de cadena sencilla (SSCP). Se observaron seis patrones de migración de SSCP distintos (A a F) con diferentes complejidades. El patrón A fue detectado en cinco departamentos en 66% de los aislados y el E solo en el departamento de Cundinamarca con una frecuencia de 0,09%. Los patrones B, C, D y F se encontraron en proporciones similares entre 13% a 5,6%, y estuvieron presentes en los cinco departamentos. La homología de nucleótidos del 75% de la secuencia del gen CP fue mayor del 99% y la tasa dN/dS (cambios no sinónimos/ cambios sinónimos) fue de 0,002. Amplicones del gen completo de CP de ocho aislados que representaban los seis patrones de SSCP se clonaron y un análisis SSCP mostró que todos tenían más de una variante. El análisis de secuencias de 35 clones confirmó la variabilidad intra aislado de PYVV. La existencia de varias variantes en un solo aislado de campo se demostró y se sugiere selección negativa de cambios de aminoácidos en al CP.
Palabras clave: PYVV, proteína de la cápside, variabilidad intra, SSCP.
The Potato yellow vein virus (PYVV) (family Closteroviridae, genus Crinivirus) is the etiologic agent of the Potato yellow vein disease (PYVD), a quarantinable disease in Europe and United States. PYVV is a re-emergent virus (Salazar, 2006) in Andean countries, infecting potato plants in Colombia, Venezuela, Perú and Ecuador (Salazar et al., 2000; USDA-APHIS 2009; EPPO, 2009). Symptoms of PYVD in potato begin with vein clearing of secondary and tertiary leaf veins, followed by a yellowing that covers the leaf lamina, resulting in the leaflet becoming bright yellow (Salazar et al., 2000). PYVD affects the weight and number of tubers produced by different varieties and Solanum species. It has been estimated that, for Solanum tuberosum subsp. andigena (STA) and Solanum phureja (SPH), losses of between 30% and 50% can occur depending upon the host, weather conditions, vector presence and other variables (Saldarriaga et al., 1988; Salazar et al., 2000;Guzmán et al., 2012; Guzmán et al., 2013).
PYVV is transmitted by the insect vector Trialeurodes vaporariorum (West) (white flies) in a semi-persistent manner by planting infected potato tubers and performing grafts with infected plants (Saldarriaga et al., 1988; Salazar et al., 2000). PYVV has a tripartite RNA (ss+) genome consisting of RNA 1 (8035 nt), RNA 2 (5399 nt) and RNA 3 (3892 nt). RNA 2 codes for the coat protein which covers most of the virion (Livieratos et al., 2004).
The population structure and genetic diversity of several plant viruses has been estimated by SSCP and nucleotide sequence analysis of genomic regions (Rubio et al., 1996; Rubio et al., 2001a; Rubio et al., 2001b; Sambade et al., 2002; Lin et al., 2003; Goszczynski and Jooste, 2002). There have been several reports on studies performed to examine the inter-isolate variability of PVYY (Offei et al., 2004; Guzmán et al., 2006; Rodríguez et al., 2010; Ayala et al., 2011). Offei et al. (2004) compared isolates obtained from 12 samples of STA and SPH collected in Colombia and one Peruvian isolate. Analysis was performed by SSCP followed by AluI enzyme restriction and the findings suggested little variability between the isolates. Screening of a larger population (250) of PYVV symptomatic Solanum spp plants obtained from three departments of Colombia using RFLP with Hinf I also demonstrated a low inter-species variability (Guzmán et al., 2006). So far, these studies have reported the variability between isolates, but, to our knowledge, there are no studies on the intra-isolate variability for PYVV. With Citrus tristeza virus (CTV) also a member of the Closteroviridae family, inter- and intra-isolate studies have been associated with different symptom expression levels, yield reduction and vector transmission (Ayllón et al., 1999; Sambade et al., 2002).
The aim of this study was to determine the inter- and intra-isolate variation of the CP gene of PYVV in samples of SPH, STA and SCH collected in five departments of Colombia. In order to study the inter-isolate variation, RT-PCR products of field samples (isolates) were subjected to SSCP and sequence analysis. To study intra-isolate variation, RT-PCR products of field isolates were cloned and the clones were further sequenced and analyzed by SSCP. These techniques were used to study subtle sequence differences between variants found in the population or within the isolates. The presence of variants with high sequence homologyin single potato isolates is reported here for the first time.
MATERIALS AND METHODS
Plant material and viral isolates. Leaf samples of 216 potato plants (100 SPH, 115 STA and 1 SCH) showing yellowing symptoms associated with PYVV were collected in the five departments of Colombia with the largest production of potato in the country: Antioquia, Boyacá, Cauca, Cundinamarca and Nariño. The plant material was collected and transported to the laboratory on ice and stored at -20°C until they were processed.
RNA extractions. Leaf samples were ground in liquid nitrogen, resuspended in extraction buffer (2% SDS Sodium dodecyl sulfate, 0.1M Tris [pH8], 2M EDTA Ethylenediaminetetraacetic acid disodium salt dihydrate) and extracted with phenol:chloroform. The supernatant was filtered through Sephadex G-50 columns as previously reported (Guzmán, 2008). The dsRNA extracts were quantified in a fluormeterQubit (Invitrogen®).
PCR. The dsRNA extracts of the field isolates were amplified by RT-PCR primers that amplify the complete CP gene of PYVV (F2Xho 5´ CTCGAGGATCCTCATGGAAATCCGATC 3´/ R3Hind 5´AAGCTTCTACTCAATAGATCCTGCTA 3´) to produce 769 pb amplicons. These primers were designed using the Peruvian accession NO_006063 reported in GenBank. The primers were designed to have XhoI and HindIII restriction sites to allow direct cloning of the amplicons. The RT-PCR reactions were performed in two steps. For reverse transcription, 200 U of MMLV (Epicentre®), 1X of RT-PCR reaction buffer, 1 mM of dNTPs, 0.4 pmol of primer R3Hind and 25 ng of RNA were used for a final volume of 12ul. The cDNA synthesis reaction was carried out at 37 °C for 60 min. For PCR, 2ul of cDNA, 1X of reaction buffer, 2.5 mM of MgCl2, 0.4 mM of dNTPs, 0.4 pmol of each primer (F2Xho/R3Hind) and 2.5 U de Taqpolimerase (BiolaseBioline®) were used for a final reaction volume of 25ul. The thermic cycle used was as follows: initial denaturation of 3 min at 94 °C, and 35 cycles for 1 min at 92 °C, 1 min at 55°C and a final extension of 10 min at 72 °C. The products were analyzed by electrophoresis using 1% agarose gel/TBE, stained with SYBR safe (Invitrogen®). In all the RT-PCR amplifications, virus free in vitro potato samples were included as negative controls, along with water as a blank. The amplicons were purified using the UltraClean PCR Clean-Up Kit (MoBio®) prior to being sent to Macrogen, Korea for sequencing.
SSCP. Purified amplicons of the field isolates or from cloned isolates were denatured and analyzed by SSCP based on the method described by Rubio et al. (1996). The DNA was separated using denaturing buffer (95% formamide, 20 mM EDTA, 0.05% xylene cyanol and 0.05% bromophenol blue) in 8% SDS-PAGE gels at 200 V for 4 h at 4 °C and stained using silver stain (Beidler et al., 1982).
Cloning of amplicons. RT-PCR products of field isolates representing different SSCP patterns were cloned in the pGEMT 3Z vector (Promega®). Escherichia coli One-Shot TOP10 (Invitrogen®) chemically competent cells were transformed according to the instructions of the manufacturer.
Sequence analysis. Field viral isolates and clones representing different SSCP patterns were sequenced in both directions (Macrogen®) using the same primers used for RT-PCR. The sequences were edited and compared with the GenBank database using the software BLAST 2.2.4. For sequence editing, EditSeq of the DNASTAR Lasergene 10 package was used. Multiple alignments were performed in Muscle V3.6 (Edgar, 2004) using the ProtTest model (Abascal et al., 2005) and the Model test algorithm (Posada and Crandall, 1998) to determine the best nucleotide and aminoacid substitution models, respectively. Nucleotide distances and the dN/dS ratio (No-synonymous /synonymous aminoacid changes) were estimated with MEGA 5 (Tamura et al., 2011). With a dN/dS<1, the selection pressure was considered negative or purifying; with a dN/dS = 1 neutral or a dN/dS>1, it was diversifying or positive. Dendograms were built in Phyml 3.0.1 (Guindon and Gascuel, 2003) by neighbor joining, taking into account the substitution model. Dendogram robustness was evaluated by 1000 bootstrap (Felsenstein, 1983). Sequences were reported in GenBank.
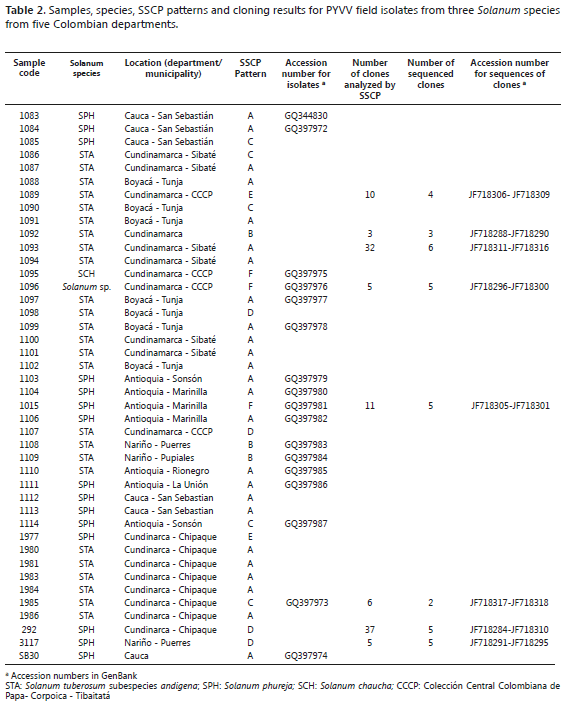
Inter- and intra-isolate variation analysis. In order to study the inter-isolate variation of PYVV CP, 216 dsRNA field samples of STA, SPH and SCH were amplified by RT-PCR and the amplicons were analyzed by sequencing and SSCP. The amplicons of 77 samples representing the six observed SSCP patterns were sequenced. To estimate the intra-isolate variability of the field isolates, that is the presence of different PYVV variants within one field isolate, the RT-PCR amplicons of the CP gene of PYVV from 5 STA and 3 SPH isolates were cloned. Between 3 and 32 clones of each isolate were subsequently analyzed by SSCP and sequencing. A total of 109 clones were analyzed by SSCP and 35 clones were sequenced.
RESULTS AND DISCUSSION
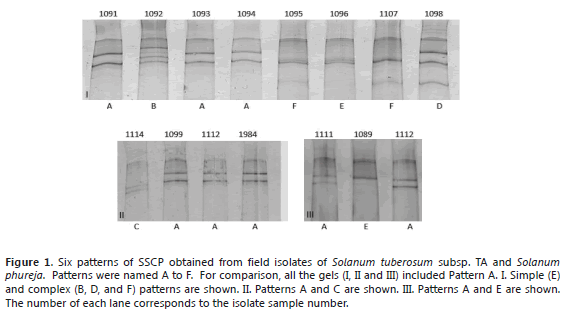
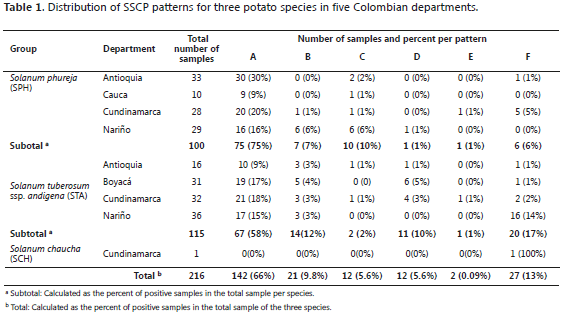
SSCP patterns from field virus isolates. Six different SSCP patterns (A to F) were found in the 216 evaluated field samples. The different patterns, simple and complex are shown in Figure 1. The SSCP patterns A, C and E are the simplest (with three clear bands), as compared to the B, D and F patterns that showed 4 to 8 bands. The six patterns were found at least once in different plants of the two major species evaluated. The most common pattern was A, present in 66% of all the isolates and in the 5 departments, 58% of STA (67 of 115 samples) and 75% of SPH (75 of 100 samples) (Table 1). Pattern E was found only in Cundinamarca with a low percent (1%), represented by one isolate from SPH and one from STA. In SPH, the second most common pattern was C (10%), followed by B (7%) and F (6%) and finally D and E in 1% of the samples. In STA, Pattern F was present in 17% of the samples, B in 12%, D in 10% and E and C in low percentages of 1% and 2%, respectively.
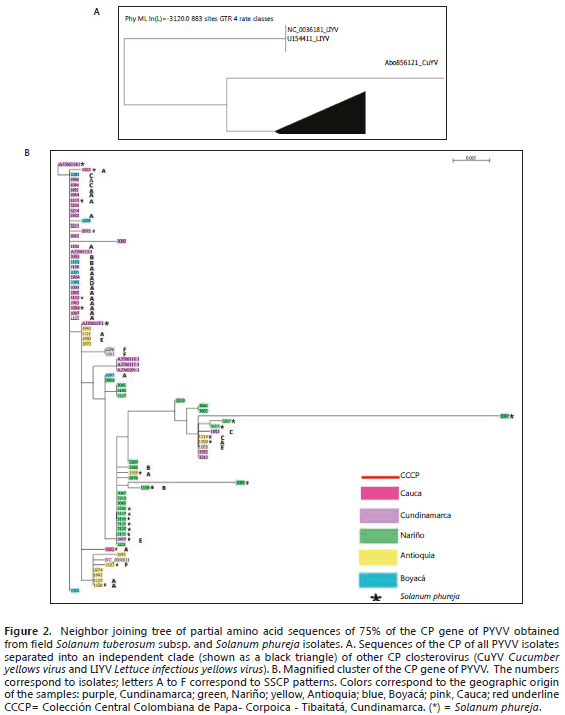
Sequence analysis of viral field isolates. CP gene sequences of 588 bp from 77 isolates, representing about 75% of the central part of the gene, were obtained. Of these, 17 were uploaded to the GenBank database (Table 2). The average nucleotide distance estimated for the 77 isolate sequences was 0.006 ±0.0029. The corresponding amino acid sequences were used to build a neighbor joining tree. The tree includes the sequence data, the geographic origin and the SSCP pattern of the selected isolates (Figure 2). All the isolates were grouped in one single clade (indicated as a black triangle) separated from the CP sequences of two closteroviruses, Lettuce infectious yellows virus (LIYV) and Cucumber yellows virus Cucumber yellows virus (CuYV) (Figure 2A). The magnification of the clade (Figure 2B) shows that the homology of these sequences was more than 99%. The sequences grouped in two big clusters, with one in the upper part of the tree composed of very similar sequences, mainly from Cundinamarca and Boyacá (neighboring departments located in the center of Colombia). The lower part of the tree contains three subclusters, in which samples from Antioquia, north of Cundinamarca and Boyacá, are spread. All the isolates from Nariño concentrate in one of these subclades. Samples of STA and SPH were present in all the clades. The different SSCP patterns did not correlate with geographic origin, the host species or any particular clade.
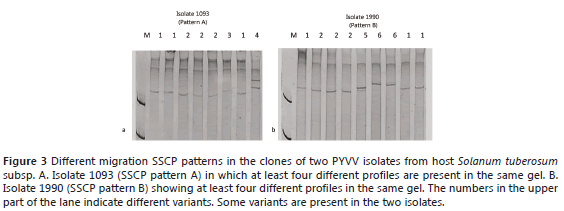
SSCP and sequencing analysis of selected clones. The SSCP of the clones confirmed that A, B, C, D, E and F were complex patterns composed of two or more CP gene variants (Figure 3). Some of the variants seemed to be present in different patterns; for example, isolates 1093 (pattern A) and 1990 (pattern B) share two different variants (named 1 and 2 in each gel) (Figure 3).
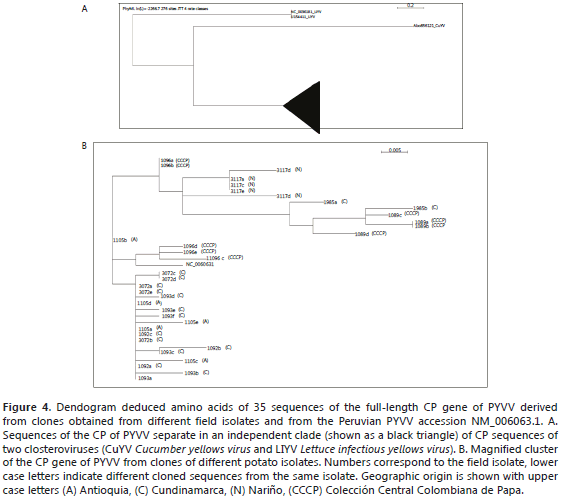
The sequence variability of the different variants existing in the particular isolates was estimated by sequencing the complete CP gene of a number of clones of each isolate. The variability of the 35 cloned sequences was 0.0091 ± 0.0030 and the dN/dS ratio was 0.0001. These sequences were used to build a dendogram (Figure 4). As with the isolates, the PYVV CP sequences separated from those of the closterovirus (CuYV and LIYV) were included in the analysis (Figure 4A). The sequences derived from the clones clustered into three groups. The sequences of clones derived from the same isolate tended to be identical or generally clustered in the same group showing low variability. Nevertheless, it was also possible to detect slight distance differences between the clones derived from the same isolate (labeled with the same isolate number and a different low case letter) (Figure 4B). The selected clones were chosen and sequenced and 35 representative sequences were reported in GenBank (Table 2). Figure 4 shows the dendogram obtained by the alignment of the 35 sequences derived from 8 cloned isolates.
In order to study the inter- and intra-variability of a population of PYVV in STA and SPH hosts, the CP gene was analyzed by SSCP and sequencing, as reported for other viruses (Rubio et al., 1996; Rubio et al., 2001a; Rubio et al., 2001b; Sambade et al., 2002; Lin et al., 2003; Goszczynski and Jooste, 2002). SSCP was performed in 216 field samples from STA, SPH and one SCH collected in five departments of Colombia. Six different SSCP patterns were detected representing more than two variants, with patterns A, C and E being less complex than B, D and F. The six SSCP patterns were found in both hosts, STA and SPH, but at different proportions (Figure 2b). Pattern A was the most common, being present in samples from all departments and detected in more than 60% of the isolates in STA and SPH. Patterns B and F were present in 9.8% and 13% of the samples, respectively; patterns C and D in 5.6% and E in 1% of the samples or less. Pattern A was more common in SPH than in STA (78% and 58%, respectively) and B and F were less common in SPH than in STA (7% and 12%, for B and 6% and 17% for F, respectively). In the SPH patterns, D and E were present in 1% of the samples, C in 10% of the samples and F in 6% of the samples. In the STA patterns, C and E were present at a low frequency (2% and 1%, respectively), D was present in 10% and F in 17% of the samples (Table 1).These results suggest that some variants are preferentially represented in one of the hosts, indicating that the selective pressure exerted by the two species may be different for these variants or that the two species may have different susceptibility to certain PYVV populations.
The SSCP patterns did not show a definitive association with the geographic origin of the SPH and STA hosts (Figure 2B); however, a number of samples from the neighboring departments of Cundinamarca and Boyacá tended to group in one cluster. The isolates collected in Antioquia spread over several clusters. Samples from Nariño concentrated in another cluster but, surprisingly, the isolates from Cauca clustered with the isolates of Cundinamarca and Boyacá but not with the samples of Nariño. These results suggest that there is movement of PYVV infected potatoes between the studied regions. There is a certification program for potatoes in Colombia; however, less than 10% of the growers use certified potato seeds, which contributes to the dispersion of viruses and other pathogens. Although the five departments in this study are located in three separate geographic regions, they are connected by roads, thus allowing for the movement of potentially infected tubers for potato seeds and commercialization. Dispersion of PYVV by the insect vector T. vaporariorum may also be involved.
One sample of a symptomatic SCH collected from the Colección Central Colombiana de Papa (CCCP Cundinamarca) was included in the analysis. Even though SCH is not an economically important potato species in Colombia, it is grown in the Andes and its response to PYVV infection is potentially important in understanding the epidemiology of this virus. The SCH sample was infected with PYVV and was shown to be the second most common SSCP for Pattern F.
Isolate sequence analysis was performed on 77 samples, representing all the SSCP patterns previously obtained. The average nucleotide variation of the CP gene (including one Peruvian and 76 Colombian isolates) was estimated at 0.006 ± 0.0029, thus confirming previous observations that have suggested low variability within this gene (Offei et al., 2004; Guzmán et al., 2006). A recent in silico analysis of the nucleotide and deduced amino acid sequence of the CP gene of PYVV confirmed that, despite the wide geographic distribution, different hosts and collecting years exist; this gene maintains a genetic similarity of between 97.1% to 100%. This indicates a high spatial and temporal genetic stability (Chaves et al., 2013). Low variability has been reported for the CP gene of other criniviruses such as Cucurbit yellow stunting disorder virus(CYSDV) and Blackberry yellow vein associated virus (BYVD), in which the mean nucleotide distance between different isolates was 0.00294 ± 0.00097 (Rubio et al., 2001a) and 6% (Poudel et al., 2012), respectively. Additionally, in order to estimate the degree and sense of selection, the dN/dS ratio was calculated. For the PYVV isolates analyzed in this study, the dN/dS ratio was 0.002, similar to 0.005 in BYVD (Poudel et al., 2012) and lower than 0.07048 as estimated for CYSDV (Rubio et al., 2001a). Negative or purifying selection has also been reported for other closteroviruses, such as Citrus tirsteza virus (CTV), in which the dN/dS between CTV isolates in different regions varied between 0.02739 and 0.56323 (Rubio et al., 2001b) and in Grapevine leafroll-associated virus 1 (GLRaV-1) in which it was 0.2521 (Alabi et al., 2011). dN/dS ratios<1 have been reported in viruses of other families (Rodríguez et al., 1989, Vives et al., 2002; Herranz et al., 2008; Komorowska et al., 2011). Recently, Martelli et al. (2012) proposed a fourth genus within the Closteroviridae family based on the divergence of the amino acid sequence of three taxonomically relevant genes as the criteria, in which there was more than 25% sequence divergence. RNA viruses have been considered rapid-evolving entities that have high mutation and recombination rates (Holland et al., 1982; Malpica et al., 2002; Worobey and Holmes, 1999) but the ability to mutate does not necessary result in high genetic variability because the genetic structure of a viral population is also shaped by other evolutionary forces, such as selection and genetic drift. According to García et al. (2001), negative selection is the most common situation for viral proteins and the evolutionary trend is towards small population diversity and genetic stability of viral genomes.
The different SSCP patterns observed in the field isolates reflect the presence of one or more variants per sample. Since PCR is performed before SSCP, any of the present variants can be amplified so the result does not necessarily reflect the actual composition of the sample. This approach allows for the detection of different variants without the need of sequencing. In SSCP, denaturing the treatment separates the two DNA strands produced by PCR and since they acquire different secondary structures, their migration pattern may differ (Rubio et al., 1996; Goszczynski and Jooste, 2002; Sambade et al., 2002). So, if only one variant is present per isolate, a pattern composed of two different bands would be expected. However, there are cases in which two different strands renature with the same secondary structure so their migration patterns could be the same. When complex patterns are observed, with more than two bands migrating differentially, the presence of several different variants may be inferred. Each pattern can consist of one or more virus variants and one variant may be present in various patterns. The PYVV field isolates studies showed that the six patterns were composed of several variants. Different SSCP patterns have been reported to display different biological activities; for example, in Beet necrotic yellow vein virus (BNYVV) (Acosta et al., 2008) and in CTV (Sambade et al., 2002). So far, there is no evidence of a differential biological behavior between different SSCP patterns of PYVV.
Evidence of different variants within one field isolate (Figure 3) was confirmed with the clone sequence analysis. For example, the clones of isolate 1096 grouped into different clusters, suggesting that different variants coexist within the same isolate, although they may differ in few nucleotides (Figure 4B). Populations of plant viruses often consist of a few genetic variants and many infrequent variants (García et al., 2001). This has been confirmed in this study for the PYVV populations in Solanum species in Colombia. For example, in Figure 3, the most common variants, labeled 1 and 2, were present in both Patterns A and B and the same variant may be present in the different isolates, such as clones 292a, 292e, 1105a, 1105d, 1092c, 292b, 1092a, 1093a, which had the same sequence (Figure 4B).
These results confirm the evolutionary tendencies observed for other members of Closteroviridae. The term quasispecies is used to describe a population of mutant viral sequences that vary around a consensus sequence (Holland et al., 1982) and it has been used to describe the intra-isolate variability observed within populations of RNA viruses (Turturo et al., 2005). However, for a population to be considered a quasispecies, the action of natural selection has to be demonstrated in the population rather than in the individual (Lauring and Andino, 2010; Holmes, 2010); a phenomenon known as the "survival of the flattest". Our results demonstrate the existence of variability, but additional tests need to be performed to demonstrate the adaptive potential of the different variants within the population.
CONCLUSIONS
Six different SSCP patterns were detected among a sample of 216 PYVV field potato isolates, suggesting inter-isolate variation. Five SSCP patterns presented complex profiles and one a simple profile. A potato isolate with a complex SSCP pattern indicates intra-isolate variation. Complex profiles were more common and widely distributed than simple profiles.
The presence of several variants in a single field isolate was confirmed by the SSCP and sequence analysis of the cloned PYVV of the field isolates. The evidence shows very similar variants within the isolates (variability of 0.0091), but not of a single homogeneous viral population.
Low variability at the nucleotide level with a dN/dS ratio of 0.002 was observed in the field isolates, suggesting negative selection against amino acid changes in the CP of PYVV.
No correlation was observed between the different SSCP patterns or sequences and the geographic origin of the samples.
ACKNOWLEDGEMENTS
The authors would like thank to the students from the Plant Virus laboratory at the Instituto de Biotecnología Universidad Nacional de Colombia. We thank the Universidad Militar Nueva Granada for providing transportation for some field visits. This work was funded by MADR - ASOHOFRUCOL (2007S4654-75, Universidad Nacional de Colombia and Universidad Militar Nueva Granada (CIAS 236). Finally we thank Dr. Helen Griffiths for reviewing this paper.
BIBLIOGRAPHY
Abascal F., R. Zardoya and D. Posada. 2005. ProtTest: selection of best- fit models of protein evolution. Bioinformatics 21(9): 2104-2105. [ Links ]
Acosta, R., M.W. Fawley and C.M. Rush. 2008. Changes in the intraisolate genetic structure of Beet necrotic yellow vein virus populations associated with plant resistance breakdown. Virology 376(1): 60-68. [ Links ]
Alabi, O.J., M. Al Rwahnih, G. Karthikeyan, S. Poojari, M. Fuchs, A. Rowhani and R.A. Naidu. 2011. Grapevine leafroll-associated virus 1 Occurs as genetically diverse populations. Phytopathology 101(12): 1446-1455. [ Links ]
Ayala, M., J.F. Gil and M. Marín. 2011. Evaluación de la variabilidad genética del PYVV en cultivos de papa de Colombia mediante secuenciación de la cápside viral. Fitopatología Colombiana 35S: 126. [ Links ]
Ayllón, M., L. Rubio, A. Moya, J. Guerri and P. Moreno. 1999. The haplotype distribution of two genes of Citrus tristeza virus is altered after host change or aphid transmission. Virology 255(1): 32-39. [ Links ]
Beidler, L.L., P.R. Hilliard and R.L. Rill. 1982. Ultrasensitive staining of nucleic acids with silver. Analytical Biochemistry 126(2): 374-380. [ Links ]
Chaves, G., M. Guzmán and L. Ortíz. 2013. Genetic structure and evidence of putative darwinian diversifying selection in the Potato yellow vein virus (PYVV). Agronomía Colombiana 31(2): 161-168. [ Links ]
Edgar, R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high through put. Nucleic Acids Research 32(5): 1792-1797. [ Links ]
European and Mediterranean Plant Protection Organization. 2009. EPPO quarantine pest. EPPO Standards. http://archives.eppo.org/EPPOStandards/PM1_GENERAL/pm102%2818%29_A1A2_2009.pdf; accessed: June 2011. [ Links ]
Felsenstein, J. 1983. Statistical inference of phylogenies. Journal of the Royal Statistical Society 146(3): 246-272. [ Links ]
García, F., A. Fraile and J.M. Malpica. 2001. Variability and genetic structure of plant virus populations. Annual Review of Phytopathology 39: 157-186. [ Links ]
Goszczynski, D.E. and A.E. Jooste. 2002. The application of single-strand conformation polymorphism (SSCP) technique for the analysis of molecular heterogeneity of grapevine virus A. Vitis 41(2): 77-82. [ Links ]
Guindon, S. and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52(5): 696-704. [ Links ]
Guzmán, M. 2008. Manual de protocolos para la detección de algunos virus que infectan la papa (Solanum spp.). Cevipapa-UNAL. Bogotá. 101 pp. [ Links ]
Guzmán, M., L. Franco, D. Rodríguez, L. Vargas and J.E. Fierro. 2012. Yield losses in Solanum tuberosum group Phureja cultivar Criolla Colombia in plants with symptoms of PYVV in field trials. American Journal Potato Research 89(6): 438-447. [ Links ]
Guzmán, M., A.K. Hernández and L. Franco. 2013. Tracking foliar symptoms caused by tuber-borne Potato yellow vein virus (PYVV) in Solanum phureja (Juz et Buk) cultivar "Criolla Colombia". American Journal Potato Research 90(3): 284-293. [ Links ]
Guzmán, M., E., Ruiz, N. Arciniegas and R. Coutts. 2006. Occurrence and variability of Potato yellow vein virus in three departments of Colombia. Journal of Phytopathology 154 (11-12): 748-750. [ Links ]
Herranz., M. C., M. Al Rwahnih, J. A. Sánchez, S.F. Elena, E. Choueiri, A. Myrta and V. Pallás. 2008. Low genetic variability in the coat and movement proteins of american plum line pattern virus isolates from different geographic origins. Archives of Virology 153(2): 367-73. [ Links ]
Holland, J., K. Spindler, F. Horodyski, E. Grabau, S. Nichol and S. Vande Pol. 1982. Rapid evolution of RNA genomes. Science 215(4540): 1577-1585. [ Links ]
Holmes. E.C. 2010. The RNA quasispecies: fact or fiction? Journal of Molecular Biology 400(3): 271-273. [ Links ]
Komorowska, B., P. Siedlecki, S. Kaczanowski, B. Hasiow-Jaroszewska and T. Malinowski. 2011. Sequence diversity and potential recombination events in the coat protein gene of Apple stem pitting virus. Virus Research 158(1-2): 263-267. [ Links ]
Lauring, A.S. and R. Andino. 2010. Quasispecies theory and the behavior of RNA viruses. Plos Pathogens 6(7): 1-8. [ Links ]
Lin, H.X., I. Rubio, A. Smythe, M. Jimenez and B.W. Falk. 2003. Genetic diversity and biological variation among California isolates of Cucumber mosaic virus. Journal of General Virology 84(1): 249-258. [ Links ]
Livieratos, I., E. Eliasco, G. Muller, R. Olsthoorn, L. Salazar, W. Pleij and R. Coutts. 2004. Analysis of the RNA of Potato yellow vein virus: evidence for a tripartite genome and conserved 3'-terminal structures among members of the genus Crinivirus. Journal of General Virolology 85(7): 2065-2075. [ Links ]
Malpica, J.M., A. Fraile, I. Moreno, C.I. Obies, J.W. Drake and F. García. 2002. The rate and character of spontaneous mutation in an RNA virus. Genetics 162(4): 1505-1511. [ Links ]
Martelli, G.P., N. Abou Ghanem-Sabanadzovic, A.A. Agranovsky, M. Al Rwahnih, V.V. Dolja, C.I. Dovas, M. Fuchs, P. Gugerli, J.S. Hu, W. Jelkmann, N.I. Katis, V.I. Maliogka, M.J. Melzer, W. Menzel, A. Minafra, M.E. Rott, A. Rowhani, S. Sabanadzovic and P. Saldarelli. 2012. Taxonomic revision of the family Closteroviridae with special reference to the Grapevine leafroll-associated members of the genus Ampelovirus and the putative species unassigned to the family. Journal of Plant Pathology 94(1): 7-19. [ Links ]
Offei, S.K., N. Arciniegas, G. Muller, M. Guzmán, L.F. Salazar and R.H. Coutts. 2004. Molecular variation of Potato yellow vein virus isolates. Archives of Virology 149(4): 821-827. [ Links ]
Posada, D. and K.A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14(9): 817-818. [ Links ]
Poudel, B., S. Sabanadzovic, J. Bujarski and I.E. Tzanetakis. 2012. Population structure of Blackberry yellow vein associated virus, an emerging crinivirus. Virus Research 169(1): 272-275. [ Links ]
Rodríguez, P., G. Chaves, L. Franco and M. Guzmán. 2010. Low molecular variability of Potato yellow vein virus (PYVV) isolates of Solanum phureja and Solanum tuberosum from Colombia. Phytopathology 100: S176. [ Links ]
Rodríguez, E., A. Moya and F. García. 1989. Variability and evolution of the plant RNA virus Pepper mild mottle virus. Journal of Virology 63(5): 2198-2203. [ Links ]
Rubio, L., Y. Abou-Jawdah, H.X. Lin and B.W. Falk. 2001a. Geographically distant isolates of the crinivirus Cucurbit yellow stunting disorder virus show very low genetic diversity in the coat protein gene. Journal of General Virology 82(4): 929-933. [ Links ]
Rubio, L., M.A. Ayllón, J. Guerri, J.H. Pappu, C.L. Niblett and P. Moreno. 1996. Differentiation of Citrus tristeza closterovirus (CTV) isolates by single-strand conformation polymorphism analysis of the coat protein gene. Annals of Applied Biology 129(3): 479-89. [ Links ]
Rubio, L., M.A. Ayllon, M. Ping Kong, A. Fernandez, M. Polek, J. Guerri, P. Moreno and B.W. Falk. 2001b. Genetic variation of Citrus tristeza virus isolates from California and Spain: Evidence for mixed infections and recombination. Journal of Virology 75(17): 8054-8062. [ Links ]
Salazar, L. 2006. Emerging and re-emerging potato diseases. Potato Research 49(1): 43-47. [ Links ]
Salazar, L., G. Muller, M. Querci, J. Zapata, and R. Owens. 2000. Potato yellow vein virus: its host range, distribution in South America and identification as a Crinivirus transmitted by Trialeurodes vaporariorum. Annals of Applied Biology 137(1): 7-19. [ Links ]
Saldarriaga A., A. Alvarez y J. Jaramillo. 1988. Efecto del amarillamiento de venas transmitido por Trialeurodes vaporariorum (Westwood) en papa. Revista Colombiana de Entomología 14(2): 3-8. [ Links ]
Sambade, A., L. Rubio, S.M. Garnsey, N. Costa, G. Müller, M. Peyrou, J. Guerri and P. Moreno. 2002. Comparison of viral RNA populations of pathogenically distinct isolates of Citrus tristeza virus: application to monitoring cross-protection. Plant Pathology 51(3): 257-265. [ Links ]
Tamura, K., D. Peterson, N. Peterson, G. Stecher, M. Nei and S. Kumar. 2011. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28(10): 2731-2739. [ Links ]
Turturo, C., P. Aldarelli, D. Yafeng, M. Digiaro, A. Minafra, V. Savino and G.P. Martelli. 2005. Genetic variability and population structure of Grapevine leafroll-associated virus 3 isolates. Journal of General Virology 86(1): 217-222. [ Links ]
USDA-APHIS. 2009. Notification of Department of Agriculture. Conditions for import of seed potatoes from the United States of America B.E. 2552. http://moacdc.thaiembdc.org/pdfs/USA_Seed%20Potatoes_ENG.pdf. 10 p.; accessed: June 2011. [ Links ]
Vives, M.C., L. Rubio, L. Galipienso, L. Navarro, P. Moreno and J. Guerri. 2002. Low genetic variation between isolates of Citrus leaf blotch virus from different host species and of different geographical origins. Journal of General Virology 83(10): 2587-2591. [ Links ]
Worobey, M. and E.C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. Journal of General Virology 80: 2535-2543. [ Links ]