The applications of enzymes for various bioproducts formulation for human needs is the driving force behind the evaluation of microorganisms for different extracellular enzymes (Kwatia and Dzogbefia, 2018). The main focus is on starch-degrading enzymes, mainly α-amylase (α-1,4 glucan-glucanohydrolase) which represent more than 60% of the enzyme market due to their usefulness in several industrial processes (Ali et al., 2017). Alpha amylase breaks down the α-1,4 glucosidic bond in starch-containing substrate resulting in monosaccharides such as glucose, short-chain dextrin, and oligosaccharides (Ali et al., 2017).
Being an extracellular enzyme amylase can be extracted from plants, animals, and certain microorganisms. However, more attention has been paid to bacterial and fungal amylases because of their thermostability, ease of cultivation, and higher productivity in industrial applications (Ali et al., 2017, Ahmad et al., 2019). Amylases from fungi are documented to be more commercially consistent than amylases from bacterial sources. They also have a highly accepted GRAS (generally recognized as safe) status. As a result, investigations have been carried out on the process parameters of promising fungal strains (Lim et al., 2020). Aspergillus spp. has been reported to possess some unique enzyme machinery with exceptional catalytic properties making them suitable candidates for industrial operations (Karim et al., 2018). For instance, A. oryzae serves as a host for heterologous protein production and possesses the capacity to produce large quantities of relevant industrial enzymes that are of great economic value. Also, Aspergillus niger highly opposes contamination and is acid-tolerant (Gurung et al., 2013).
Solid substrate fermentation (SSF) has been considered a promising technique for the production of enzymes due to its economic and engineering advantages (Razdan and Kocher, 2018). In recent years, the SSF process has been developed and extensively used for the synthesis of bio-products, especially when it involves fungi (Kwatia and Dzogbefia, 2018). SSF cultivated microbes have tight contact with insoluble substrates letting a higher nutrient utilization from them and achieving high enzyme yields during fermentation. Numerous advantages are attributed to SSF these include higher volumetric production, utilization of simple fermentation media, require less water, generate, fewer effluents, no laborious control of process parameters, bacterial contamination is reduced, and requires less capital involvement for further processing (Ahmad et al., 2019).
With the high demand for novel amylases, getting a cheap, rich, and the readily available suitable substrate is key to meeting the high demand with excellent catalytic efficiency. Hence, cheaper and readily available starchy foods such as cereals (rice, corn, wheat, and millet) and tubers (yam, cassava, potatoes, and cocoyam), could be adopted as an additional use in starch enzymatic breakdown to produce reducing sugars. The enzymatic is inexpensive in terms of energy consumption and the use of a simpler process than the conventional that utilizes pregelatinized starch as substrate (Kwatia and Dzogbefia, 2018).
Purification is an important aspect after enzyme production to obtain a purified component from the crude extract. Without purification, enzyme activities and associated proteins might be difficult to characterize accurately due to the impurities in the crude solution, resulting in unreliable data and information (Lim et al., 2020). Searching for a novel fungal strain with high catalytic properties at a low cost, the present study isolated and cultivated an amylolytic fungus using an economic substrate for amylases production, purification, and characterization. The fungal strain purified amylases demonstrated some biochemical properties making them promising candidates to use in different industries processing starch.
MATERIALS AND METHODS
Collection of samples
Grain samples namely: rice (Oryza sativa), wheat (Triticum aestivum), corn (Zea mays), and millet (Pennisetum glaucum) were purchased at Oja-oba market, Akure, Nigeria. Debris and unwanted particles were sorted out from the grains. They were washed, ground into fine particles, packaged in transparent plastic bags, labeled, and transported to the laboratory for further microbial and biochemical analysis.
Fungal isolation and identification
Fungi were isolated from fermented cereals using potato dextrose agar (PDA) by standard pour plate techniques and incubated at 30 °C. The fungal colonies were sub-cultured to purify the isolates. The isolates were prepared on slides and stained with lactophenol cotton blue to identify their morphology and compare them with a standard fungal atlas (Pitt and Hocking, 2009).
Qualitative screening of isolates for amylase production
Alpha-amylase producing isolates were screened on starch-agar medium (soluble starch 5 g L-1; peptone 5 g L-1; yeast extract 5 g L-1; MgSO4·7H2O 0.5 g L-1; FeSO4·7H2O 0.01 g L-1; NaCl 0.01 g L-1; agar 15 g L-1 plates. The isolate was inoculated at the center of the plates, at an appropriate temperature and then was stained with Lugol solution. The colonies forming the largest halo zone were selected for further studies (Yalcin and Çorbaci, 2013).
Genetic identification
The genetic identification of the isolates was performed by extracting DNA following the standard technique of Xian et al. (2015). Then, using PCR the partial internal transcribed spacer (ITS) region genes were amplified with the corresponding primer sets that target ITS1 (5'-TCC GTA GGT GAA CCT GCG G-3') and ITS4 (5'-TCC TCC CC GCT TAT TGA TAT GC-3') (Xian et al., 2015). The amplification protocol adopted was: 40 PCR cycles with denaturation at 95 °C for 2 min, annealing at 58 °C for 2 min, and extension at 72 °C for 2 min, with the final extension for 10 min. The sequences of the amplicon were compared with the sequences in the GenBank using a BLASTN search and aligned with related species according to Xian et al. (2015).
Fungal inoculum
Spore suspension of the amylase-producing fungal isolates was prepared by scrapping off fungal spores which were dissolved in 40 mL of distilled water until the final volume of 60 mL and vortexed. The SSF production medium was inoculated with 2 mL fungal spore suspension (Kwatia et al., 2017). The mineral salt medium (MSM) contains 0.1 g KH2PO4, 0.25 g NaCl, 0.01 g MgSO4·7H2O, 0.01 g FeSO4.7H2O, and 0.10 g yeast extract in 100 mL of distilled water.
Amylase production in SSF
To choose the best substrate in the SSF for amylase production, commercial starch was substituted with corn, rice, wheat, and millet as selected organic substrates. The organic substrate was used singly; 100% of corn (C); rice (R); wheat (W); millet (M) or in a mixture at different ratios: 1:1 of corn and rice (CR); corn and wheat (CW); corn and millet (CM); rice and wheat (RW); rice and millet (RM); wheat and millet (WM); 1:2:1 corn, rice, and wheat (CRW); rice, corn, and millet (RCM); millet, wheat and rice (MWR); wheat, millet, and corn (WMC); 1:1:1:1 corn, rice, wheat, and millet (CRWM). 4 g of each combination was dissolved in 10 mL of MSM and sterilized at 121 °C for 15 min. At room temperature, the dissolved substrate was inoculated with 2 mL of the fungal spore suspension, thoroughly mixed, and incubated aerobically at 30 °C for 96 h (Kwatia et al., 2017).
Enzyme extraction
The extracellular crude enzyme was extracted from the fermentation medium using 0.05 M citrate phosphate buffer (pH 6.0). 50 mL of the buffer was dispensed into the 4 days old enzyme production medium, homogenized, and kept in a rotary shaker (250 rpm) at 30 °C for 30 min. The resulting mixture was filtrated using a cheesecloth and the filtrate was centrifuged (3600 xg, 15 min at 4 °C). The clear crude extract had crude enzymes for amylolytic activity evaluation (Kwatia and Dzogbefia, 2018). The composite substrate with the highest α-amylase production was considered a potential substrate.
Amylase assay
The amylase activity was determined following the protocol of Ahmed et al. (2020). A mixture of 0.5 mL of crude extract and 0.5 mL of soluble starch was incubated at 40 °C in a water bath for 15 min, the reaction was stopped with 1 mL of 3, 5 dinitrosalicylic acid, boiled for 5 min and cooled to measure the reducing sugars and estimate the enzymatic activity according to the method of Miller (1959). The commercial substrate, i.e., soluble starch was prepared by dissolving 1% (w/v) of the substrate in 0.05 M citrate phosphate buffer (pH 6.0). The absorbance was taken at 540 nm against a substrate blank using the UV Spectrophotometer (Axiom 721 vis spectrophotometer). The quantity of reducing sugars released from the enzyme-catalyzed starch was estimated using a glucose standard curve. A unit of amylase activity was indicated as the amount of enzyme needed to liberate 1 μmol of reducing sugars (maltose/glucose) per minute under standard assay protocols (Ahmed et al., 2020).
Determination of protein concentration in the crude extract
The protein contents in the crude extract were evaluated by the Bradford method (1976). Different concentrations (0.1-1.0 mg mL-1) of Bovine Serum Albumin were prepared and used as a standard. Finally, the absorbances of the samples were measured at 595 nm.
Crude amylase purification and molecular weight
The crude enzyme was purified by three purification steps: ammonium sulphate precipitation, ion exchange, and gel filtration chromatography (Abdulaal, 2018). After purification, the biochemical characteristics of the purified enzyme, kinetic parameters, and molecular weight were determined.
Ammonium sulphate precipitation
Solid ammonium sulphate was continuously and gently dissolved in crude enzyme solution until 60% of saturation. Allowing the salt precipitation in the mixture at 4 °C for 12 h, the saturated solution was centrifuged for 20 min at 14,000 rpm to obtain the pelletized proteins, which were dissolved in 10 mL of 0.05 M citrate phosphate buffer (pH 6.0). Furthermore, the protein was dialyzed in the same buffer for 96 h (Hassan et al., 2018).
Dialysis
The precipitated protein was aseptically poured into a dialysis bag and dialyzed in the same buffer for 96 h in refrigeration until releasing the sulphate from the solution, replacing the buffer in the bag embedded. After dialysis, the protein content and α-amylase activity were subsequently determined (Hassan et al., 2018).
Ion-exchange chromatography
The dialysate free of salt was loaded on a DEAE Sephadex a-50 column (2.5×40 cm), previously treated with 0.05 M citrate phosphate buffer pH 6.0. The protein was eluted using the same buffer at a flow rate of 2 mL min-1. The unbound inactive protein was eluted from the column, followed by the elution of the bound proteins by a linear salt gradient (0.1-1.0 M NaCl). The protein content eluted was determined by reading the absorbance at 280 nm and the α-amylase activity was assayed following the prescribed protocol. The fractions showing enzyme activities were concentrated with 4 M sucrose solution (Hassan et al., 2018).
Gel filtration
The concentrated enzyme was further purified using a Sephadex G-100 column (2.5×75 cm) previously treated with 0.05 M citrate phosphate buffer pH 6.0. The protein was eluted using the same buffer at a flow rate of 20 mL h-1. The protein content of each fraction was monitored by reading the absorbance at a wavelength of 280 nm and α-amylase activity was assayed as previously described. The pooled fractions were stored at 4 °C and used for biochemical characterization (Hassan et al., 2018).
Biochemical characterization of the purified α-amylase
The biochemical properties of the purified enzyme and kinetic parameters, such as Km and Vmax, were studied. The influence of temperature and pH on the activity and stability of purified α-amylase and the effect of metal ions and inhibitory agents on enzyme activity were also determined.
Influence of temperature and pH on the catalytic activity and thermal stability of purified α-amylase
To determine the optimal temperature for α-amylase activity, equal volumes of enzyme and substrate were incubated at different temperatures, from 40 to 90 °C. The thermal stability of the enzyme was evaluated by taking samples every 30 min for 2 h, and the residual activity was evaluated following standard assay protocol (Abdulaal, 2018).
The influence of pH on the catalytic activity of purified α-amylase was determined by incubating at different pH in 50 mM buffer system: glycine-HCl buffer (pH 3.0), sodium acetate buffer (pH 4.0-5.0), phosphate buffer (pH 6.0-7.0), Tris- HCl buffer (pH 8.0-9.0) and glycine-NaOH (pH 10.0-12.0). The stability of the purified enzyme was monitored by incubating in the appropriate buffer solutions for 2 h and taking an aliquot for enzymatic assay every 30 min (Abdulaal, 2018).
Influence of metal ions and inhibitors on the catalytic activity of purified α-amylase
The influence of different concentrations of metal ions and enzyme inhibitors (5 and 10 mM) on the catalytic properties of the enzyme was determined by incubating the mixture enzyme/substrate with the metal ions (Ca2+, Cu2+, Mg2+, Mn2+, Ni2+, Hg2+, Na+, Zn2+, K+, and K3Fe (CN) or the inhibitors (EDTA and urea). After incubation, the enzymatic activity was determined following standard assay procedures (Abdulaal, 2018).
Estimation of kinetic parameters of the enzyme
The initial rate of the amylase-substrate reaction was measured using soluble starch (1-10 mg mL-1) as substrate. The kinetic parameters (Km and Vmax) of the purified enzyme were estimated from the double reciprocal plot of Lineweaver and Burk (Abdulaal, 2018).
Determination of molecular weight of purified α-amylase
The homogeneity and subunit molecular weight of the purified α-amylase was determined by 10% gel sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a wide range protein molecular weight marker as standards from 11 to 180 kDa and stained with Coomassie brilliant blue (Abdulaal, 2018).
RESULTS AND DISCUSSION
Presumptive identities of fungal isolates from corn, rice, wheat, and millet cereal flour
The isolated fungi from the cereal samples of wheat, rice, corn, and millet were tentatively identified as Aspergillus niger, A. flavus, Rhizopus stolonifer, Fusarium poae, Penicillium chrysogenum, and Cladosporium sp. From all the samples, Aspergillus spp. occurs frequently followed by Penicillium spp. The result is similar to that of Hussain et al. (2018) who also identified five fungal genera from cereals of wheat, barley, corn, and rice and documented Aspergillus as the most predominant. The predominance of Aspergillus on fermented cereals could be that they are degraders and common spoilage organisms of carbohydrate-containing (Hussain et al., 2018).
In addition, the genetic identity of the best α-amylase producer resulted in the fungus as A. flavus with 97% similarity. The amylase activities of the fungal isolates on different organic substrates are presented in Figure 1.
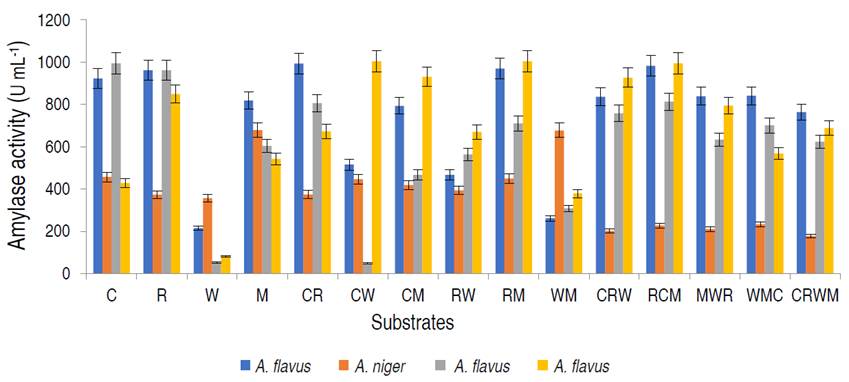
Figure 1 Amylase activity of the amylolytic fungi on different organic substrates viz; 100% corn (C); rice (R); wheat (W); millet (M); 1:1 corn and rice (CR); corn and wheat (CW); corn and millet (CM); rice and wheat (RW); rice and millet (RM); wheat and millet (WM); 1:2:1 corn, rice and wheat (CRW); rice, corn and millet (RCM); millet, wheat and rice (MWR); wheat, millet and corn (WMC); 1:1:1:1 corn, rice, wheat and millet (CRWM).
Aspergillus flavus isolated from millet had the highest amylase activity of 1.004 U mL-1 on the combination of rice and millet (RM), A. flavus from rice was observed to have the highest amylase activity on rice and millet, with values of 0.814 U mL-1, while A. niger from corn had the best activity on millet only. Based on α-amylase activity, A. flavus from millet was selected for purification and characterization studies. The production of α-amylase on different substrates from fungal strains using SSF has been studied. Kwatia et al. (2017) and Sadhasivam et al. (2018) produced amylase from A. niger and A. luchuensis BS1 by SSF using agro-residues. Abdulaal (2018) and Balakrishnan et al. (2021) also separately produced α-amylase from Trichoderma pseudokoningi and A. oryzae via SSF using orange peels and edible oil cakes, respectively.
In SSF, the selection of a suitable culture medium and a suitable microbial strain are key factors for the production of α-amylases (Razdan and Kocher, 2018). This study revealed appreciable amylase production by A. flavus in the composition of rice and millet. This study is in agreement with Ali et al. (2017) who documented higher amylase by A. flavus on mandarin peels. Oppose to this study, Oshoma et al. (2017) found maximum α-amylase yield in A. niger using different agricultural wastes via SSF. These variations might be due to the differences in physiological status, and genetic makeup of the organisms leading to varied abilities in utilizing the solid substrates for α-amylase production (Badejo et al., 2021).
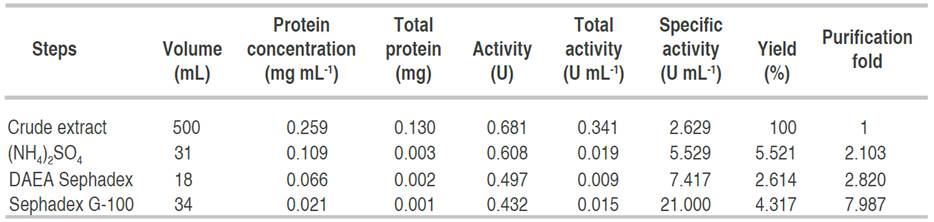
Purification summary of amylase
The elution profile of α-amylase from A. flavus on the ion exchange column is presented in Figure 2.
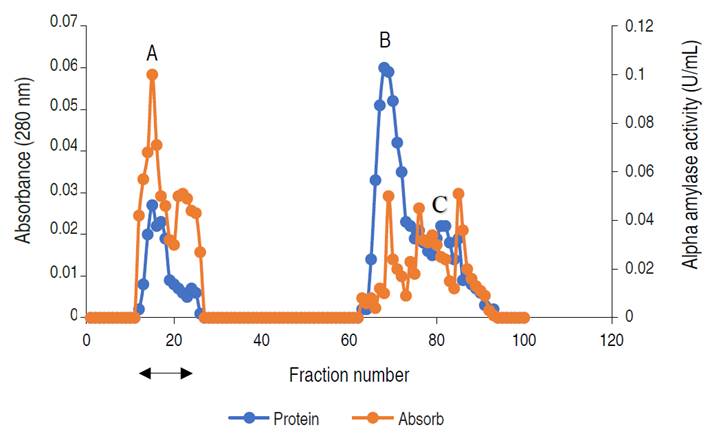
Figure 2 Chromatogram of α-amylase produced by A. flavus on ion-exchange column (2.5×40 cm) of DEAE Sephadex.
The profile revealed three (3) activity peaks labeled as A, B, and C. Each peak was pooled together to form the three fractions. The fractions forming peak A had an activity of 0.497 U mL-1, peak B of 0.169 U mL-1, and peak C constitutes the lowest activity of 0.024 U mL-1. The major active peak A was pooled together and was further subjected to purification by gel chromatography. The elution profile of α-amylase from A. flavus via gel filtration is shown in Figure 3.
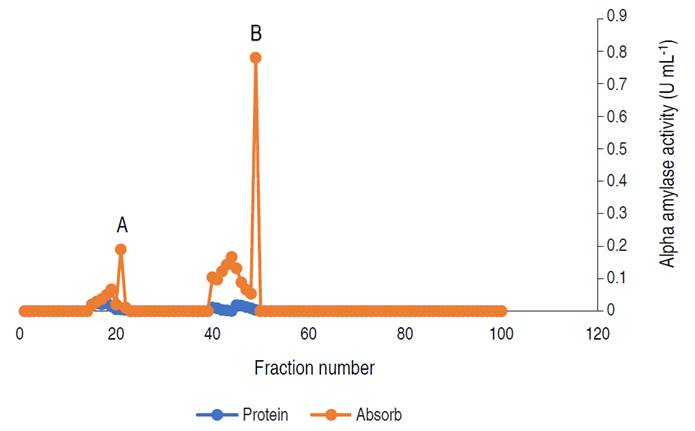
Figure 3 Elution profile of α-amylase via gel filtration with two activity peaks. Pooled fractions were represented with black arrow.
The profile revealed two (2) activity peaks (A and B). Peak B had the highest activity hence, fractions from peak B were pooled together and used for the characterization.
Table 1 has a summary of the processes and the techniques used for the obtaining homogeneity of the enzyme. Alpha amylase from A. flavus was purified to homogeneity with a purification yield of 4.3% and a fold of 7.9. Similar to this result, Nithya et al. (2017) recorded a purification fold of 7.06, while Hassan et al. (2018) and Kwatia et al. (2017) reported a lower purification fold of 4.78 and 4.48, respectively. Higher purification folds of 18 and 16.6% were documented by Abdulaal (2018) and Gbenga et al. (2017) for purified α-amylase from different fungal isolates. The differences in the purification yield and fold by different authors is due to the type of resins used and other factors such as buffers and purification conditions (Badejo et al., 2021). Fold purification indicates the purification status of the enzyme; an increase in fold purification reveals an increase in the purity of the enzyme subjected to purification when compared with the crude enzyme (Badejo et al., 2021).
Physicochemical properties of a purified amylase
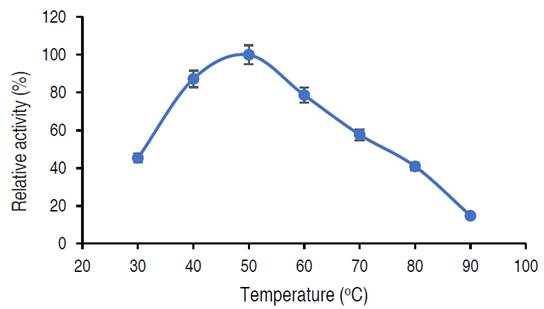
Figure 4 shows the catalytic activity of the purified amylase. The highest temperature was considered as 100% and the rest of the temperature was estimated relative to the maximum activity. The relative activity of purified α-amylase increased from approximately 45% at 30 °C and reaches its peak at 50 °C with the value of 100% relative activity, beyond 50 °C, a steady decrease in relative activity was observed, at 90 °C, the activity decreased to 14%.
In this current study, the activity of purified α-amylase reached optimum at 50 °C. Abdulaal (2018) also recorded maximum α-amylase from Trichoderma sp. at 50 °C. Similarly, El-Sayed et al. (2019) also documented optimum amylase activity from Bacillus mojavensis at 55 °C. Higher and lower temperatures than those obtained in this study had been recorded from other studies; Hassan et al. (2018) showed optimum amylase activity from E. coli at 30 °C, while Wang et al. (2018) showed that the amylase activity from Pseudoalteromonas sp. was 25 °C. Sadhasivam et al. (2018), Trabelsi et al. (2019) and Wang et al. (2018) in separate studies documented optimal activities at 60 °C for purified α-amylase from A. luchuensis, B. subtilis and Thermomyces dupontii (expressed in Komagataella phaffii, respectively. The decrease in α-amylase activity beyond 50 °C might be connected to the disruption of various bonds in the amylase (Wang et al., 2018). While the variations observed in the optimal temperatures from different studies could be due to the source of the microbial enzyme and differences in the genetic makeup of the organisms (Badejo et al., 2021).
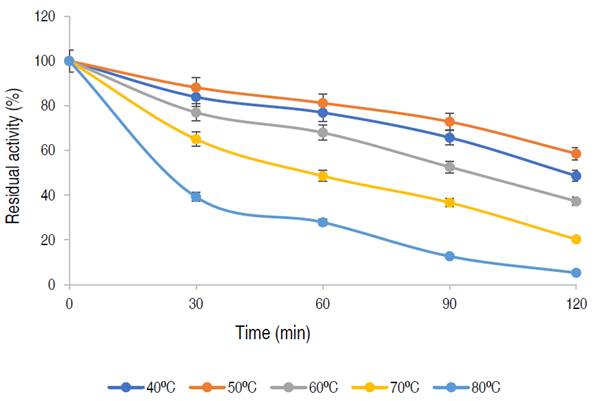
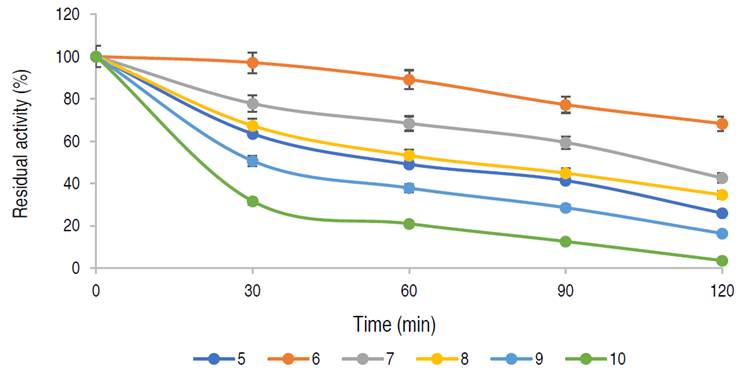
Thermal stability of the purified α-amylase
The infuence of temperature on the stability of the enzyme is shown in Figure 5.
The enzyme has a residual activity of approximately 81% after 1 h at 50 oC and 58% of its activity was retained after 2 h, while 76%, 67%, and 48% of residual activities were retained at 40 oC, 60 oC, and 70 oC, respectively after 1 h of incubation. The purified α-amylase from this study was stable and retained 81%, 67% and 48% of its activity at 50 °C, 60 °C and 70 °C for up to 60 min respectively. Karim et al. (2018) similarly documented thermo-stable α-amylase from A. flavus NSH9 at 50 °C, retaining 87% of its residual activity after 60 min of incubation. The decline in enzyme activity beyond the period of stability could be due to hydrolysis of peptide chain or aggregation, thus causing abnormality in the conformation of the enzyme (Karim et al., 2018).
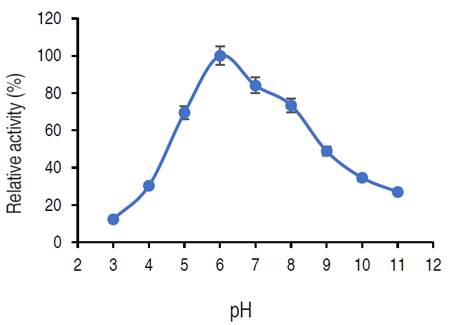
Influence of pH on purified α-amylase activity
The influence of pH on purified α-amylase is presented in Figure 6.
The relative activity of purified amylase increased from pH 3.0 and reached its optimum at pH 6.0 and then decreased. At pH 8.0 and 9.0, the relative activities were approximately 73% and 48% respectively.
The purified α-amylase from this study was active both in acidic and alkaline regions (5.0 to 8.0), where an optimum pH was recorded at pH 6.0. The purified α-amylase from different fungi have pH optimum ranged from acidic (3.0) to alkaline region (11.0) (Abdulaal, 2018). Different studies have reported various pH optimal for purified α-amylase. Doss and Anad (2012) reported 6.0 as the best pH for optimum α-amylase activity from A. flavipes and A. wentii. Trabelsi et al. (2019) similarly recorded a pH of 6.0 for maximum purified α-amylase activity from different sources. On contrary, a pH of 7.0 was optimum for amylase from A. nidulans and A. versicolor (Sethi et al., 2016). Hammami et al. (2018) and Priyadarshini and Ray (2019) reported an optimum α-amylase from Bacillus mojavensis SA and a Bacillus sp. at pH 9.0. The variations in the optimal pH observed in different studies could occur as a result of the microbial origin of the enzyme as well as modification of the genetic make-up of the isolate (Badejo et al., 2021). Also, the activity of the purified α-amylase over a wide range of pH shows the versatility of the α-amylase for several processes. The decline in α-amylase activity at pH 4 and 10 might be due to the denaturation of essential co-factors or dissociation of the enzyme structure (Badejo et al., 2021).
pH stability of the purified α-amylase
The influence of pH on the stability of the purified α-amylase is presented in Figure 7. The purified α-amylase was 100% relatively stable at pH 6.0 for 30 min, and after 2 h of incubation, 68% residual activity was retained. The enzyme retained more than 40% of its residual activity after 90 min at pH 8.0. The stability of this enzyme fulfills the requirement for its application in many industrial operations especially for starch processing.
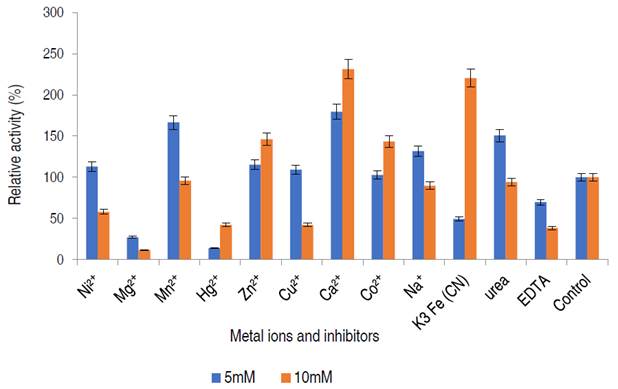
Influence of metal ions and inhibitors on α-amylase activity
The effect of different metal ions and enzyme inhibitors on the purified α-amylase from A. flavus is presented in Figure 8.
The α-amylase activity was stimulated in the presence of Ni2+, Mn2+, Cu2+, Ca2+, Na+, Zn2+, Co2+ and urea at 5 mM, while Hg2+ and EDTA mildly inhibited α-amylase activity at concentration dependent manner.
In this study, the purified enzyme was stimulated by some salts, notably calcium ion. Many of these metal ions are known to enhance enzyme activity, some metal ions act as cofactors increasing the activity of amylase for starch hydrolysis (Lim et al., 2020). From this study, the addition of Ca2+, Co2+ and Zn2+ positively modulated purified α-amylase at both concentrations of 5 and 10 mM, amylase activity was only activated by Na+, Cu2+, Ni2+ and Mn2+ at 5 mM while the enzyme activity was inhibited in the presence of EDTA, and Hg2+. A similar observation was documented by Kwatia and Dzobgefia (2018) and Aladejana et al. (2020) when α-amylase from B. subtilis was incubated with Ca2+. Also, Bashir et al. (2014) documented stimulation of α-amylase from B. lichiniformis on the addition of Zn2+, Co2+, and Mn2+. Ca2+ activated α-amylase suggest that calcium ion might act as an activator and stabilizer of α-amylase and that requirement for Ca2+ differs for different α-amylases (Maalej et al., 2021).
In contrary with this study, the inhibitory action of Ca2+ at 10 mM against purified amylase was documented by Babu and Satyanarayana (1993). EDTA had an inhibitory effect on the α-amylase activity. Chelating agents such as EDTA are known to inhibit enzymes by either detaching metal ions from the enzyme or by binding to the enzyme to form a ligand (Aladejana et al., 2020).
The findings of this study suggests that the enzyme from A. flavus is a metalloenzyme and that the enzyme contains metallic ions which are removed by the chelating agent, forming an active complex with EDTA and consequent loss of activity (Aladejana et al., 2020). In general, variations from different studies occurs because the metal-binding ability of α-amylases, were found to be dependent on the enzyme sources (Maalej et al., 2021).
Kinetic parameters of purified α-amylase
The kinetics parameters, maximum velocity (Vmax) and Michaelis-Menten constant Km are showed in Figure 9.
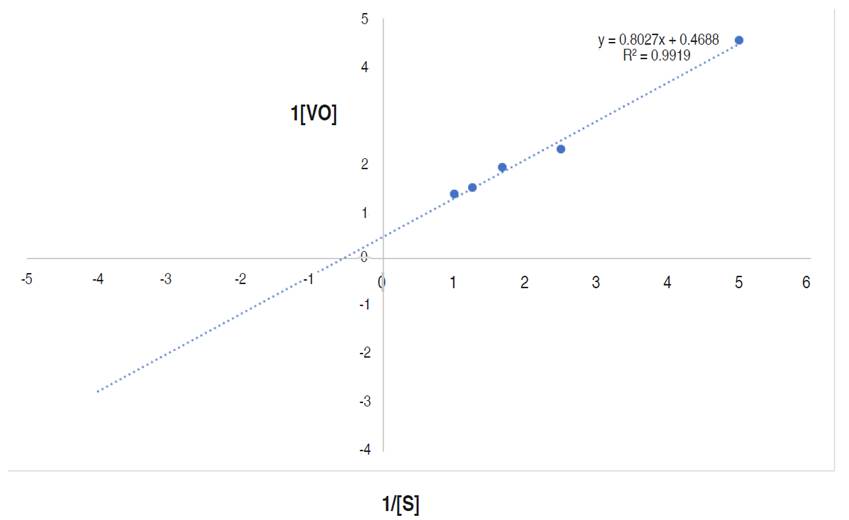
Figure 9 Line weaver-Burk plot of reaction velocity against substrate concentration for purified α-amylase using starch as substrate.
This research documented the apparent Km and Vmax of the purified α-amylase to be 1.71 mg mL-1 and 2,133 µmol min mL-1 respectively. Higher Km and Vmax of α-amylase from Bacillus sp. MB6 were documented as 5.45 mg mL-1 and 24.15 μmol min mL-1 (Paul et al., 2017). Abdulaal (2018) also reported the Km and Vmax values for purified α-amylase from Trichoderma pseudokoningii to be 4.0 mg mL-1 and 0.74 μmol min mL-1. Km measures the affinity of the enzyme for substrates i.e., the enzyme substrates complex, while the Vmax of an enzyme is the measure of the maximum rate of reaction. The Km of an enzyme, relative to the concentration of its substrate under normal assay conditions allows prediction of whether or not the rate of product formation, will be affected by substrate concentration (Badejo et al., 2021). From this study, the higher Vmax and lower Km values suggest a reasonable bonding between the substrate and enzyme. It also confirms the efficiency of the purified α-amylase for diverse industrial applications (Badejo et al., 2021) such as in food industries.
Molecular weight estimation
The most active fraction from the purified amylase subjected to SDS-PAGE for molecular weight determination was estimated to be 45 kDa. SDS-PAGE is considered to determine the molecular mass of the purified enzyme as well as a method to indicate the purity of an enzyme.
Maalej et al. (2021) similarly documented 45 kDa in the study of a novel digestive α-amylase from Blue Crab. Xian et al. (2015) reported higher molecular weight of 70 kDa from purified α-amylase obtained from Talaromyces pinophilus. The variations in molecular weights obtained from different organisms might be related to isozymic properties as well as types of amino acid residues of the enzyme (Maalej et al., 2021).
CONCLUSION
The fungus A. flavus isolated from fermented cereals cultivated on low-cost feedstock via SSF demonstrated α-amylase activity with excellent biochemical properties. The biochemical properties of the purified α-amylase showed that it is acidophilic and thermotolerant with a high affinity for the substrate. These are the properties required in a wide range of industries. Therefore, the combination of rice and millet as substrate serves as a good medium for higher amylase production for various industrial applications.