The specialized milk production systems with bovines constantly face the presence of pathogens that affect the sanitary conditions and the economy of herds. One of these pathogens is bovine leukemia virus (BLV), whose target cells are B lymphocytes, causing enzootic bovine leukemia (EBL), which affects the immune system of animals causing immunosuppression and allowing entry to other pathogens (Bartlett et al. 2020; Marawan et al. 2021). Most animals are asymptomatic to the disease; only 30% of infected animals develop persistent lymphocytosis and only 1-5% develop lymphomas (Barez et al. 2015). In addition to affecting the health of animals, it generates a detriment in the quantity of milk produced and the compositional and sanitary quality of the milk, having adverse effects on the economy of dairy herds (Marawan et al. 2021; Úsuga-Monroy et al. 2018a).
The BLV belongs to the Retroviridae family; these viruses are characterized by transcribing their genome from RNA to DNA through the reverse transcriptase enzyme and inserting themselves into the host cell genome (Polat et al. 2017). When this occurs, the virus is renamed provirus; infections of this type are permanent. BLV transmission occurs through contact with fluids that have the presence of infected cells (blood, colostrum, and milk) from an infected animal to another healthy one, for which good management practices in production systems are vital to prevent virus transmission (Bartlett et al. 2014; Ruiz et al. 2018). It has also been found that it can be transmitted in the womb and at the time of delivery (Kuczewski et al. 2021).
The viral genome consists of four primordial genes that code for structural proteins and enzymes (env, gag, pol, and pro). In addition, it has two regulatory genes (tax and rex) and the R3 and R4 genes that code for accessory proteins, and a part that encodes microRNAs. The envelope gene (env) encodes proteins gp30 and gp51, which play a fundamental role in the infection process, are the recognition center of B lymphocytes and modulate the immune response, so changes in their structure can affect the infective capacity of the virus (Canova et al. 2021; de Brogniez et al. 2016). Therefore, it is a highly conserved genome region and is the most used worldwide for virus detection and genetic research (Blazhko et al. 2020).
In 2016, the molecular prevalence of BLV in animals of the Holstein breed of the department of Antioquia was evaluated, finding 47.6% of positivity (Úsuga-Monroy et al. 2018c). Two years later, in a study carried out in six regions of the country, 73% of positivity was obtained in Antioquia, but only one municipality was evaluated (Corredor-Figueroa et al. 2020). Thus, this is the first extensive study that covers different breeds and geographical regions of specialized dairy in Antioquia. Although various studies have shown the implications of BLV in milk production systems (Bartlett et al. 2014; Úsuga-Monroy et al. 2018a), the virus is not included among the mandatory control diseases in the country by the Colombian Agricultural Institute (Instituto Colombiano Agropecuario - ICA), although, since 2015 and according to Resolution No. 003714 of 2015 (Instituto Colombiano Agropecuario 2015), it is mandatory to declare if its presence is found.
The lack of control mechanisms and knowledge regarding this pathogen by producers have contributed to the increase in the number of positive animals, facilitating the circulation of BLV within dairy herds. Milk production is a sensitive sector of the national primary economy, and the department of Antioquia is one of the leading milk producers in the country, with three regions (north, east, and Valle de Aburrá) where this activity prevails for the peasant economy. With this context in mind, the aim of this study is to evaluate the prevalence of BLV in the main dairy breeds and their crosses in the specialized dairy herds of this department.
MATERIALS AND METHODS
Experimental design
The study population consisted of 224,714 mechanical and traditional specialized dairy milking cows from Valle de Aburrá and the north, and east regions of Antioquia, Colombia, the largest milk-producing regions in the department (Secretaría de Agricultura y Desarrollo Rural, 2019). The sample size was calculated from the study population, with an expected molecular prevalence of 47% (Úsuga-Monroy et al. 2018c), a relative error of 10% (i.e., an absolute error of 4.4%), and a design effect of 1.2, obtaining a sample size of 575 bovines that were selected in 16 municipalities of the three regions mentioned above. The number of bovines sampled per herd was proportional to the sample size and the total number of bovines in the herd; these were selected randomly. Each herd was given a list of bovines that were selected.
Ethical aspects: Ethics Committee
This research is part of the macroproject "Bovine leukemia virus in dairies of Antioquia: Evaluation of the zoonotic potential and the effect on reproductive performance", endorsed by the Institutional Committee for the Care and Use of Animals (CICUA) of Universidad Nacional de Colombia "020-2020". For the development of the project, no permits or licenses were required. However, in the execution of the project, the legislation and other current regulations pertaining to it in terms of ethics, environmental regulations, or access to genetic resources were complied with. The permits for taking blood samples from the animals and obtaining the required information from each herd were processed with the owners of the herds included in the research, who signed a consent.
Blood sampling
Blood samples were taken from the middle coccygeal vein with an 18G needle with a vacutainer vacuum system (VACUETTE®) and tubes with EDTA as an anticoagulant to obtain the DNA of the nucleated peripheral blood cells. All samples were homogenized by inversion and transferred to the Animal Biotechnology Laboratory of Universidad Nacional de Colombia, Medellin, under refrigeration conditions (4 °C).
DNA extraction
Blood samples with anticoagulant were centrifuged to obtain the layer of white cells from which the DNA was extracted using the salting out technique (Miller et al. 1988). The DNA obtained was resuspended in TE 1X pH=8.0 buffer (1 M Tris HCl and 0.5 M EDTA) and stored at 4 °C until analysis. The sample was run on a 1% agarose gel stained with SYBR-safe to verify the integrity of the extracted DNA. Extracted DNA was quantified in a spectrophotometer (NanoDrop2000®, Massachusetts, United States) at 260 and 280 nm optical densities to verify its concentration as well as the quality through the 260:280 ratio. In addition, a PCR for a bovine constitutive gene (Toll-like receptor 6) was performed with random samples to verify that there were no inhibitors in the samples.
Molecular analysis for the detection of BLV in cattle Controls
As a positive control for the PCR tests, a mixture of DNA from bovines previously diagnosed positive serologically by the ELISA technique and by PCR for the env gene by the BIOGEM group was used. Negative controls were performed with a reaction in the absence of DNA and with DNA from a bovine previously diagnosed by serology and molecularly as negative for BLV infection.
Nested PCR of the env gene
The PCR to detect the BLV env gene was performed in a final volume of 25 μL with 150 ng of DNA, 0.2 mM of dNTPs, 1 IU of Taq polymerase, 1X of buffer, and 1.5 μL of 10 mM of each envFw and envRv oligonucleotide (Fechner et al. 1996) for the first reaction, and envFW2 and envRV2 (Fechner et al. 1996) for the second reaction (Table 1). The reaction conditions for the two PCR cycles were 5 min at 94 °C, 40 cycles of 30 s at 94 °C, 30 s at 60 °C, 1 min at 72 °C, and 5 min at 72 °C. At the end of the second reaction, a 444 bp fragment of the BLV provirus env gene was obtained in the positive animals. Agarose gel electrophoresis was performed to observe the PCR results in which the positive animals for the BLV exhibited a band at the height of 444 pb.
Statistical analysis
The results of the PCR were integrated into an Excel database with the information of the sampled animals (breed, age, parity, region and herd), and descriptive statistics were used to analyze the results. The qualitative traits were summarized in absolute and relative frequencies, estimating 95% confidence intervals. Quantitative variables were summarized using the mean and standard deviation or median with quartiles, depending on data normality. The Chi-square test (X2) was performed to determine the possible association between breed and the presence or absence of the virus. To establish the association between breed and the presence of BLV, breeds and breed types were grouped into: Holstein x Crossbreed: where crossbred can be Viking Red, Ayrshire, BON, Brown Swiss, Normande, Angus; Other purebred: where it can be: Jersey, Ayrshire, BON, Brown Swiss, Viking Red and Other crossbreeds: where there were more than three crossbreds. And they were analyzed through an Odds ratio (OR) by means of a 2x2 contingency table. It was determined that if the OR is more than one, the breed or crossbreed has a low risk of being positive for BLV and if the OR is less than one, the breed or crossbreed has a high risk of being positive for BLV. The significance of the data was accepted as P<0.05. OR analysis was performed using Epi Info™ v7.2 CDC software.
RESULTS AND DISCUSSION
The fragment amplified in the PCR corresponds to the region that codes for the surface protein gp51, vital in the infectivity cycle of the virus since it is responsible for the recognition of the receptor and the binding of the virus to the host cell. Therefore, it is decisive to know the range of virus hosts (Gao et al. 2020). The primers selected to carry out the nested PCR of the env gene have been used in various diagnostic and genotyping studies of the virus since 1996 due to the low genetic variability of this region concerning other genome regions (Fechner et al. 1996; Polat et al. 2017).
The results obtained in this study for the 575 bovines belonging to the specialized dairy in Antioquia show that the prevalence of bovine leukemia virus in the department is 17.04% (Table 2), i.e., lower than the results obtained in previous studies (Corredor-Figueroa et al. 2020; Ortiz et al. 2016; Úsuga-Monroy et al. 2018c). This result may be due to the fact that this is the first extensive study carried out in the region where cattle of different racial components (Holstein, Jersey, Viking Red, Ayrshire, and their Jerhol crosses and trihybrids, among others), age groups (calves, heifers, and cows in production) and geographical regions of the department were sampled. Furthermore, this research contrasts with previous studies where only the population of one breed (Úsuga-Monroy et al. 2018c), a municipality, or an age group were sampled (Corredor-Figueroa et al. 2020; Ortiz et al. 2016).
The low prevalence may also be the result of improved sanitary measures in specialized dairy herds in the department, the implementation of good livestock practices, and biosecurity protocols in the herds that prevent the transmission of the virus, such as the use of one needle per animal, effective as a control strategy for the BLV (Kuczewski et al. 2019). In Colombia, since 2012, the payment of raw milk to producers dependent on compositional and hygienic factors has been implemented, finding that this measure has been effective in stimulating progress in production systems (Carulla and Ortega 2016) and is directly related to the improvement of the sanitary scheme of the production systems and the general management of the animals in it. Nevertheless, the prevalence in herds was relatively high (71.7%) in this study, indicating that in more than half of the productive systems, there is at least one infected animal, existing the risk of an increase in the number of infected animals by contagion. Therefore, it is necessary to evaluate what factors can increase or decrease the risks of spreading the disease in the dairy systems of the department. It is well known that practices such as using shared needles for the application of medicines are one of the main risk factors for the infection of bovines in herds (Kuczewski et al. 2021; Ortiz et al. 2016). Moreover, it is essential to transmit information to producers, creating control strategies to contribute to the progress of this productive sector.
In the department of Antioquia, the main breeds found in specialized dairies are Holstein and Jersey. Crosses between them and with other reference breeds are also widely used when analyzing prevalence data according to racial components (Table 3). It can be inferred that this factor may be influencing the results obtained since the categories Holstein x (Cross) and other crossbreeds are associated with an increased risk of being infected with BLV. There is evidence that genetic factors can make animals susceptible or resistant to diseases. An example is the Colombian Blanco Orejinegro (BON) breed, where some studies have determined the resistance/susceptibility of these individuals to some viral infections such as foot-and-mouth disease and vesicular stomatitis (Lopez-Herrera et al. 2000, 2002).
In the case of EBL, it has been found that the molecular prevalence of the virus is lower in BON compared to data from pure breeds (Herrera et al. 2011; Úsuga-Monroy et al. 2018b). The heritability of this susceptibility factor to the bovine leukemia virus infection among Holstein and Jersey cattle populations has also been estimated at 8%, indicating that genetics plays an important role in the incidence of BLV in different cattle breeds (Marawan et al. 2021). In order to analyze the breed factor, several categories were combined to determine whether or not the racial factor has an influence on BLV infection. These results can be a precedent for later studies where this hypothesis was evaluated in depth. It should be noted that the results obtained on the prevalence of BLV in the specialized dairy in Antioquia for the different racial components are not sufficient to make comparative statements between susceptibility/resistance to BLV infection due to the number of individuals for some breeds. However, to establish the association between the breed and the presence of BLV, the following groupings were made: Holstein x Cross, others purebreds and other crossbreeds and compared with the Holstein breed (Table 3). It was identified that the racial component (Holstein x Cross and other crossbreeds) does influence the probability of not having BLV infection (P<0.05) compared to the Holstein breed. Therefore, these results show a trend that generates the hypothesis that there may be breeds with greater resistance to infection by the virus in the Antioquia dairy, however, our data do not allow us to establish individually which breed or breeds are more resistant to BLV infection.
It has been found that in the bovine major histocompatibility complex (BoLA), with a fundamental role in the presentation of antigens and the immune response of the organism against infection, there is a genetic region (BoLA-DRB3) that influences the development of BLV infection (Lo et al. 2020; Notsu et al. 2021). Most cattle with specific mutations in the allele (DRB3*009:02) maintain undetectable proviral loads and are positively related to the development of lymphomas (Lo et al. 2020). Identifying these genetic variations associated with resistance to BLV infection helps develop control programs based on the genetic selection of animals with these genes (Andoh et al. 2021). Studies recommend the evaluation of this factor in BLV positive and negative bovines from dairies in Antioquia to determine if it is the determining factor for the low molecular prevalence in the population studied.
It has been found that the association between BoLA-DRB3 gene polymorphisms and BLV infection varies between breeds (Hernandez et al. 2018; Lo et al. 2021a), supporting the hypothesis that some breeds are more susceptible than others to infection with BLV. Moreover, environmental factors also relate to the susceptibility/resistance that cattle may have (Andoh et al. 2021). Therefore, more in-depth studies are needed to test this hypothesis as, so far, only animals heterozygous for BoLA-DRB3 have been shown to have advantages against BLV infection regardless of their breed (Hernandez et al. 2018; Lo et al. 2021b).
Another factor that can be decisive in the prevalence results of BLV in this study is the age of the sampled animals. Other studies have found that the number of infected animals increases as age increases (Murakami et al. 2011). Calves are less likely to be infected, as the main transmission route of the virus is iatrogenic in production systems (Ruiz et al. 2018). It is until after the first calving that prevalence rates increase due to increased exposure to the virus. Nevertheless, the presence of the virus has been found in calves because there is a risk of transmission during pregnancy and birth and later with milk consumption (Marawan et al. 2021; Ruiz et al. 2018). It should also be considered that the onset of the persistent lymphocytosis stage in infected cattle generally occurs before 5 years of age, and after that, between 5 and 8 years of age, it passes to the lymphoma stage (Zyrianova and Kovalchuk 2019). Therefore, many adult cattle individuals may be positive for viral infection and show positive serological tests but are negative for the molecular detection test by PCR since BLV-positive cells may be sequestered in lymphomas and not be circulating in the blood.
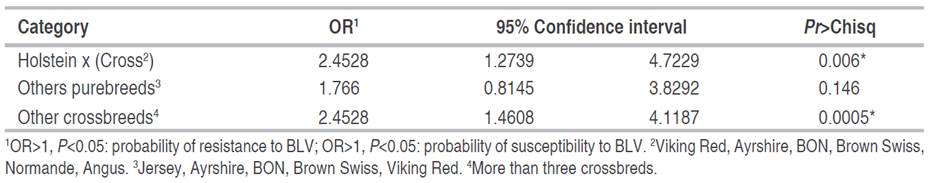
When detailing the seropositivity of the study samples by the number of calvings (Figure 1), it is observed that as the number of calvings increases, the prevalence rate also increases, reaching the highest percentage in the group of cows from four to six calvings (20.7%). Further, the positivity rate decreases in the interval of more than seven calvings, possibly due to the sequestration of BLV-positive cells in internal tissues. On the other hand, it is known that most of the cattle in specialized dairies throughout the country are discarded before reaching seven calvings due to the reduction in productive traits. It is then possible that the lower prevalence in this range of calvings is because the animals that continue in the system after seven calvings have good health conditions, and their productivity has been efficient throughout their lives. Knowing the effects of BLV infection on productive and reproductive characteristics, the possibility that these animals were not infected with BLV during their productive life and, therefore, have not been discarded should be considered.
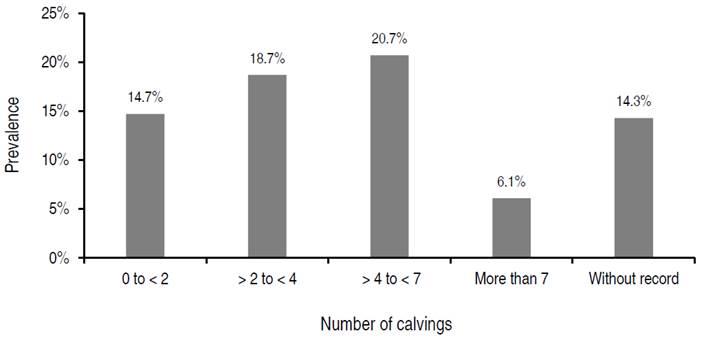
Figure 1 Results of molecular prevalence to BLV for bovines with a different number of calvings in the specialized dairy in the department of Antioquia. Group 0<2 calvings includes animals from three months of age until the first calving.
Detailing the analysis of seropositivity in the three geographical areas where the study was carried out (Figure 2), the area with the highest prevalence was Valle de Aburrá, with 21.05% positivity to BLV. This area has a lower percentage of livestock production systems dedicated to dairy than the north region of Antioquia, in which the lowest molecular prevalence was found (15.65%).
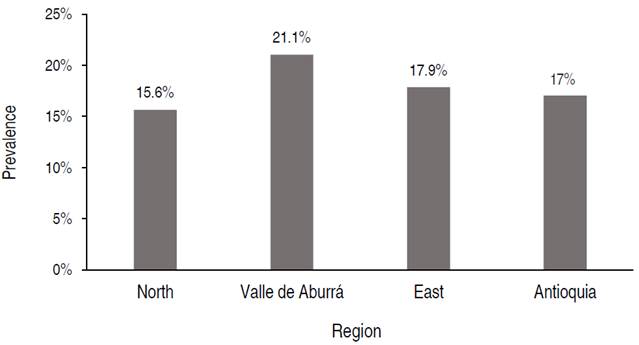
Figure 2 Results of molecular prevalence to BLV for the north (15.6%), east (17.9%), and Valle de Aburrá (21.1%) regions in the department of Antioquia, Colombia
The analysis of the results obtained in the 16 municipalities evaluated in three regions (Figure 3) shows that there is variability in these, finding values from 7.1% (Carmen de Viboral from the eastern region) to 61.5% (Girardota from the Valle de Aburrá region). Nonetheless, it should be noted that these values cannot be compared because the experimental design was based on the population of each region and not on the one at the municipality level. However, it is a result that must be considered when proposing future studies. In Valle de Aburrá, the municipality with the highest prevalence was Girardota (Figure 3A). Of the three municipalities evaluated in the region, Medellín has the lowest prevalence despite being the one with the largest bovine population included in the study for this region. The second is the east region (Figure 3B), with the municipality of Santuario exhibiting the highest prevalence (50%) with respect to the other four evaluated. Conversely, the municipality with the lowest BLV seropositivity, Carmen de Viboral (7.1%), is also found in this municipality. The north region (Figure 3C) was the one where a higher number of municipalities were evaluated (eight) since it is the one with the most specialized dairy, having most a low prevalence rate (between 7.7 and 33.3%) compared to the municipalities of other regions. On average, seropositivity for this region is 17%, which is the same value that the molecular prevalence of BLV had for the entire department. These results show a variation between the different geographical areas of the department, and future studies would be necessary to evaluate this and the possible reasons for these differences.
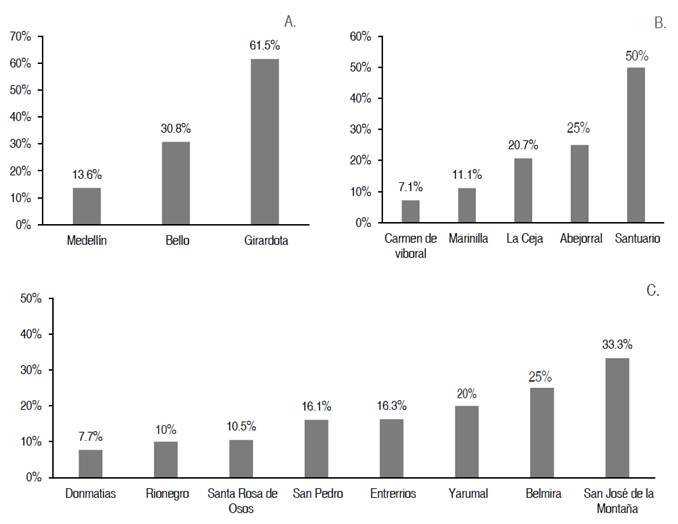
Figure 3 Molecular prevalence of bovine leukemia virus in municipalities of the department of Antioquia A. Molecular prevalence in the municipalities of Valle de Aburrá, B. Molecular prevalence in the municipalities of the east region, and C. Molecular prevalence in the municipalities of the north region.
CONCLUSIONS
The bovine leukemia virus is a pathogen found in most of the specialized dairy systems of the department of Antioquia, generating economic losses to producers. Thus, it is necessary to implement measures to control and prevent this disease in the territory. The results of this study contribute to the creation of BLV control strategies in the specialized dairy of Antioquia since differences in molecular positivity to BLV were detected by the number of calvings (age of the cows), racial component, herd, region, and municipality. It is necessary to identify factors contributing to reducing the presence of the virus in the territory to optimize this agricultural sector of the economy in the department. Thus, future studies are necessary to identify in greater detail the behavior of this pathogen under the different conditions that exist in the dairy production systems of Antioquia, as well as the influence that the different racial components may have.