Bananas are important global horticultural commodities ranked fourth after wheat, rice and corn; with over 130 countries in the tropical and subtropical regions of the world cultivating it for staple food (Perrier et al. 2011). Southeast Asia, including Indonesia, is considered the diversity and domestication center of bananas. There are at least 325 known banana cultivars in Southeast Asia and 200 of them are found in Indonesia (Hapsari et al. 2015). Therefore, Indonesia was categorized as ranked third as one of the world largest banana producers in 2019 with a contribution of 6.05% of the total world production (FAO 2020). Nevertheless, banana bunchy top disease (BBTD) caused by a banana bunchy top virus (BBTV) has become the most serious and destructive disease to threat the banana production in Indonesia (Hapsari and Masrum 2012).
BBTD is a viral disease, named after the symptoms, where the infected plants are stunted and have "bunchy" leaves at the top (Qazi 2016). When plants are infected early, they do not bear fruit, and when they are infected later, the fruits are poor and unmarketable. This virus infects all members of the Musaceae family, with several related families as alternate hosts such as Araceae, Heliconiaceae, Strelitzeaceae, and Zingiberaceae (Pinili et al. 2012). The banana aphid, Pentalonia nigronervosa (Family Aphididae; Order Hemiptera), was found to be a vector for transmitting and spreading the disease wider. Unlike fungal diseases, BBTV cannot be transmitted mechanically through garden tool use (Niyongere et al. 2012).
The virus is identified to the genus Babuvirus in the family Nanoviridae. It is an icosahedral virus consisting of six circular single-stranded DNA genome components i.e., DNA-R, DNA-U3, DNA-S, DNA-M, DNA-C and DNA-N (previously named DNA-1 to DNA-6, respectively) (Elayabalan et al. 2015). The sequences length of each genome component is approximately 1 to 1.1 Kb, and the transcripts have been mapped (Abdel-Salam et al. 2012). DNA-R encodes a master replication-associated protein (rep), DNA-S a capsid protein (cp), DNA-M a movement protein (mp), DNA-C a cell-cycle link protein (Clink), and DNA-N a nuclear shuttle protein (nsp) gene. Meanwhile, the function of DNA-U3 is still unknown (Stainton et al. 2015).
Understanding the origin of the BBTV is an important issue, and it is necessary to mitigate and control the future spreads. The origin of the BBTV is thought to come from the area of origin of bananas, namely Southeast Asia, including Indonesia (Perrier et al. 2011). Subsequently BBTV spread to a number of countries, including the Old World (36 countries in Africa, Asia, and Oceania) and there is no record of BBTV being found in the New World except in Hawaii and the United States (Qazi 2016). BBTV can be categorized into two groups based on their phylogenetic, including the Pacific Indian Ocean group and the Asian group (Stainton et al. 2015). However, due to human evolution and intermediary processes, new variants of BBTV possibly have emerged (Yu et al. 2019).
This study was aimed to confirm the BBTV infection on asymptomatic wild bananas Musa acuminata Colla varieties compared to symptomatic banana cultivars from East Java, Indonesia, through PCR-based detection using specific primers of BBTV DNA-R. The master replication initiation protein encoded by the DNA-R component of the BBTV has been successfully used for virus detection in both wild and cultivated bananas of Asian countries group (Chiaki et al. 2015; Rahayuniati et al. 2021a). Previous studies reported that wild bananas have important traits such as a relatively harsh environment and diseases resistant's, including BBTV (Hapsari and Masrum 2012), whereas banana cultivars with one or two B genomes (ABB and AAB) tend to be more tolerant to BBTD than AA and AAA (Hapsari and Masrum 2012). In addition, this study also aims to analyze molecular characteristic and phylogenetic of the BBTV DNA-R sequences with other isolates already reported in GenBank, particularly from Indonesia and Asian countries. The results of this study are expected to be useful as basic information for further mitigation and evaluation of BBTV resistant bananas.
MATERIALS AND METHODS
Plant materials
Plant materials examined in this study were four specimens of banana collection of Purwodadi Botanic Garden in Pasuruan, East Java, Indonesia. It comprised of two BBTV asymptomatic wild bananas and two BBTV symptomatic banana cultivars (Table 1, Figure 1). Asymptomatic wild bananas have a hypothesis as species that are resistant to BBTV, while symptomatic banana cultivars have a hypothesis as positive control and species that are susceptible to BBTV. For molecular study samples, young and curled leaves were taken from the living plants, stored in a cool box, and immediately transferred to the laboratory for DNA isolation.
DNA isolation and amplification
Total genomic DNA was extracted using Wizard® Genomic DNA Purification Kit from Promega. The DNA isolation steps follow the guidelines for plants. The amplification was using specific BBTV DNA-R primers (Wickramaarachchi et al. 2016) i.e., BBTV DNA-1F: 5'-GGA AGA AGC CTC TCA TCT GCT TCA GAG AGC-3' and BBTV DNA-1R: 5'-CAG GCG CAC ACC TTG AGA AAC GAA AGG GAA-3'. PCR amplification process was carried out in 30 µL of the total volume consisting of 3 µL (25 ng of DNA sample), 3 µL primer (10 µM), 6 µL of nuclease-free water and 15 µL of DreamTaq DNA. The PCR thermal protocol was carried out for 35 cycles consisting of initial denaturation of 94 °C for 3 min, denaturation of 94 °C for 30 seconds, annealing of 47 °C for 60 seconds, an extension of 68 °C for 30 seconds, and final extension at 72 °C for 10 min. Visualization of the amplified product was carried out by electrophoresis on 1.5% agarose gel with a voltage of 100 volts for 45 min. The PCR products were then purified and sequenced at 1st BASE Laboratories Sdn Bhd, Malaysia by Sanger dideoxy sequencing technology using an ABI PRISM 3730xl.
Data analysis
BBTV DNA-R sequences were evaluated using SeqScanner v1.0 software. The sequences identification and confirmation were performed using the nucleotide BLAST search in GenBank database (Altschul et al. 1990). Open reading frames (ORFs) and translation of ORFs was determined using ORF Finder server tool (Wheeler et al. 2003). All predicted ORF were aligned using protein BLAST search in GenBank database (Altschul et al. 1997) to identify the protein. Data sequences polymorphisms were analyzed using DnaSP6.
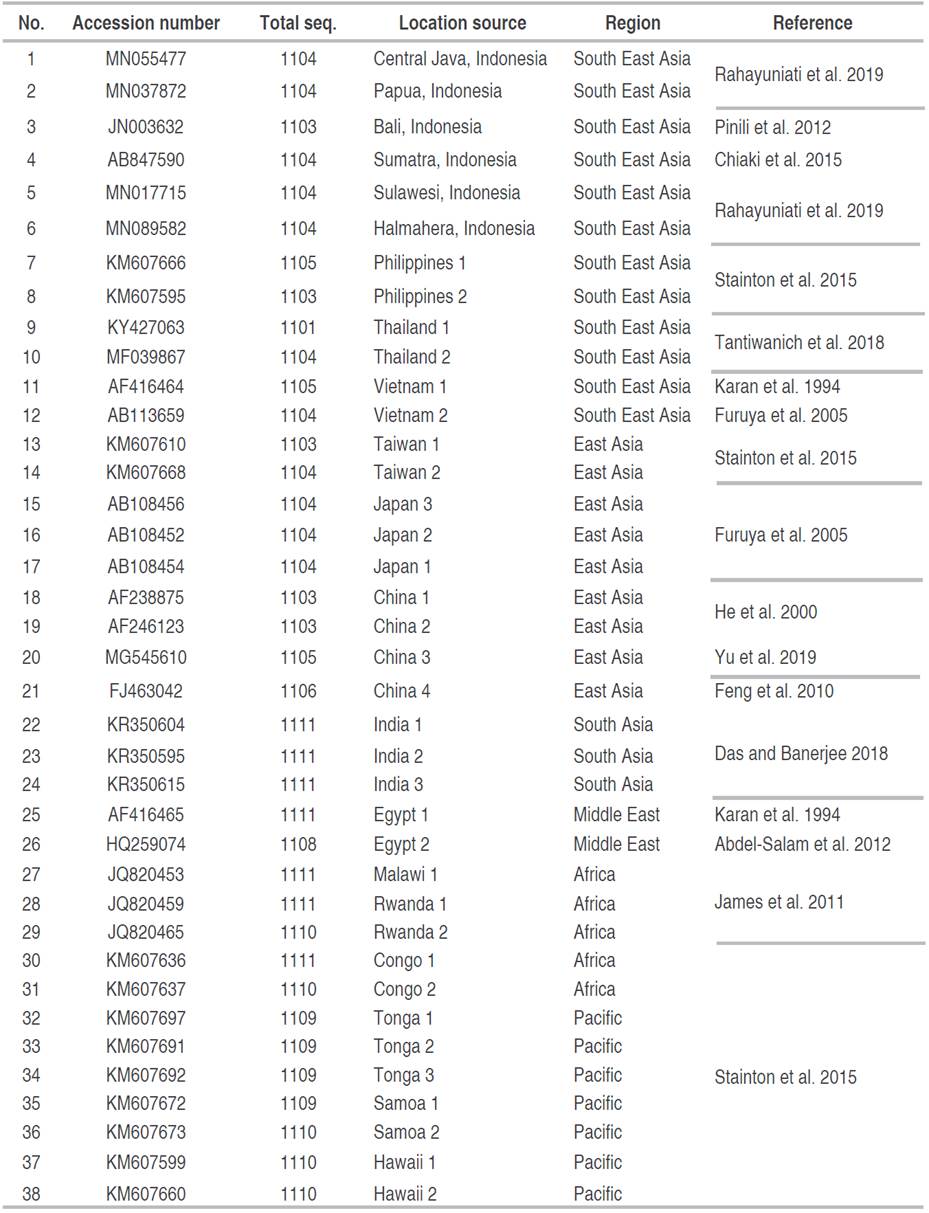
For further phylogenetic study, homolog sequences of BBTV DNA-R across the world were retrieved from GenBank database (Table 2). The all-DNA sequences were then aligned using ClustalW in MEGA7 software (Kumar et al. 2016), followed by manual adjustment and converted to suitable formats (FASTA). The evolutionary history was inferred using the Kimura two-parameter model (Kimura 1980) and Neighbor-Joining method (Saitou and Nei 1987) with 1,000 bootstrap replications.
RESULTS AND DISCUSSION
PCR amplification and sequence analysis of BBTV DNA-R
The wild bananas examined produce normal fruits and do not show any obvious bunchy top symptoms (Figure 1A and B). Meanwhile, the banana cultivars examined showed severe bunchy top infections (Figure 1C and D). This condition was in agreement with the previous study by Hapsari and Masrum (2012) which indicated that wild banana species were more resistant to BBTV than banana cultivars. Furthermore, the genotypes of banana cultivars are correlated with resistance to BBTV where banana cultivars with one or two B genomes tend to be more tolerant. In spite of that, it seems that cultivars with AAA genomes are not all equally susceptible to BBTV. Ngatat et al. (2017) stated that Gros Michel (AAA, Cavendish sub-group) exhibits resistance to the BBTV under both experimental inoculation and field conditions.
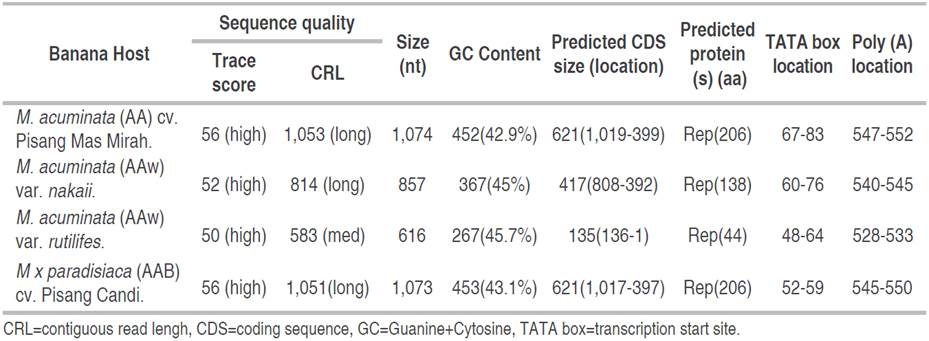
BBTV DNA-R primers were successfully amplified in all isolates, both BBTV symptomatic and asymptomatic. This indicates that BBTV DNA-R components were detected in all isolates examined, both symptomatic and asymptomatic. Furthermore, sequencing results showed high-quality value DNA sequences (Table 1) with medium and long contiguous read length (CRL) and high trace score value. Interestingly, the amplicons show different sequence lengths and contrasting with expected amplicons size (1.0 to 1.1 kb) (Stainton et al. 2015). The sizes of the BBTV DNA-R components in this study were varied from 616 to 1,074 bases (Table 3).
The asymptomatic wild bananas showed shorter or partial length amplicons than symptomatic banana cultivars that showed full length (Table 3). Nevertheless, all BBTV DNA-R detected in this study were still predicted as Rep Protein (Table 3). This is presumable to be related to the resistance mechanism of wild bananas to BBTV. Since, only part of DNA-R fragments were found (not full length) so that viral master replication protein transcription failed to form and infect the plants (asymptomatic).
The characteristics of BBTV DNA-R from banana isolates in this study were differ with banana isolates from Sri Lanka as described by Wickramaarachchi et al. (2016) in the length of predicted coding sequence (CDS), protein, TATA box and polyadenylation or poly (A) location; even though the same primer was used. Furthermore, it was also differing in the absence of the conserved regions defined as stem-loop common region (CR-SL) and major common region (CR-M). In general, each component of the BBTV genome (except DNA-R) has one big (monocistronic) transcriptional active open reading frame (ORF) and two conserved regions: TATA box at 3' of the stem-loop, CR-SL and CR-M, and poly (A) (Islam et al. 2010). CR-SL is common region with the conserved nonanucleotide (TATTATTAC), an origin of virion DNA replication (Wickramaarachchi et al. 2016). Whilst the CR-M is the second common region as the binding site for ssDNA primers and a prime the synthesis of transcriptionally active dsDNA (Das and Banerjee 2018). The absence of CR-SL and CR-M in this study may involve to the BBTV resistance mechanism in bananas.
BBTV DNA-R polymorphism
The nucleotide composition of BBTV DNA-R isolates were low in GC content, averaging 42.9% (Table 3). It is relevant to the expectation that single-stranded DNA viruses should display, on average, lower G and C frequencies compared to double stranded since ss genomes are prone to mutations toward A and T/U (Simon et al. 2021). Furthermore, sequences alignment of four isolates showed that 607 sites were conserved (56.47%), nine sites were polymorphic due to mutations, and 616 were missing data or gaps or deletions. The polymorphic sites comprised of eight singleton variables and one parsimony informative. All of the singleton variables were found in wild M. acuminata var. rutilifes, which mostly due to transversion (Ti/Tv=0.33). The singleton variables found in site positions of 20(C→A), 558(G→C), 596(T→A), 610(T→C), 613(T→C), 620(A→C), 624(A→C), and 634(A→T). The parsimony-informative were found in M. acuminata var. rutilifes and Pisang Mas Mirah at site number 351, due to transversion (A→T).
BBTV DNA-R sequences alignment and comparison of two asymptomatic wild bananas showed large gaps or missing data (241 sites; 28.82%) and moderately conserved (607 sites; 70.83%). About nine mutations were identified mostly due to transversion (Ti/Tv)=0.29) and no parsimony informatives. The singleton variables found in site positions of 13(C→A), 344(A→T), 551(G→C), 589(T→A), 603(T→C), 606(T→C), 613(A→C), 617(A→C), and 627(A→T). Meanwhile, BBTV DNA-R of two symptomatic banana cultivars showing highly conserved region up to 1,055 sites (98.14%) and only four missing data. There were 16 polymorphic sites identified mostly due to transversion (Ti/Tv=0.78) and no parsimony informatives. The singleton variables found in site positions of 4(C→A), 6(T→C), 7(C→G), 10(T→C), 12(C→T), 13(A→C), 351(T→A), 1032(T→G), 1058(C→T), 1059(T→C), 1063(G→A), 1065(T→G),1071(G→C), 1073(T→C), 1074(G→T), and 1075(A→G). These results indicated that BBTV DNA-R in wild bananas are more variables than that of banana cultivars, which thought to be the reason they show resistance (asymptomatic).
Phylogenetic tree of BBTV DNA-R isolates from East Java with others
Multiple sequences alignment and phylogenetic analysis of isolates from East Java (this study) were conducted with other 38 accessions of homologs BBTV DNA-R sequences from Asia, Africa, and the Pacific countries (Table 2). The total aligned and selected BBTV DNA-R sequences length of 42 accessions were 630 bp. The sequences were considered highly polymorphic reached 64.60% (407 sites), only about 196 positions (31.11%) were identified as conserved regions, and 27 gaps (4.29%). Furthermore, the nucleotide composition were low in GC content (43.21%).
The bootstrap consensus of Neighbour-Joining phylogenetic analysis of BBTV DNA-R in this study delineates into two large groups in relevant to many previous studies. It was separated into the Pacific Indian Ocean group/PIO (Group I) and the Asian group (Group II) with moderate to strong bootstrap support (Figure 2). BBTV DNA-R isolates from the Pacific, South Asia, Middle East and Africa countries were clustered in Pacific Indian Ocean Group and has wider distribution than Asian group that was comprised of BBTV DNA-R from East Asia and South East Asia countries. The transfer of infected banana material is thought to be the primary cause of the PIO group's wider geographic dispersion than the Asian group. Differential adaptation of PIO and Asian groups of BBTV isolates is suggested for different banana species (Wickramaarachchi et al. 2016), but this hypothesis cannot be proven because of the lack of an infectivity assay system.
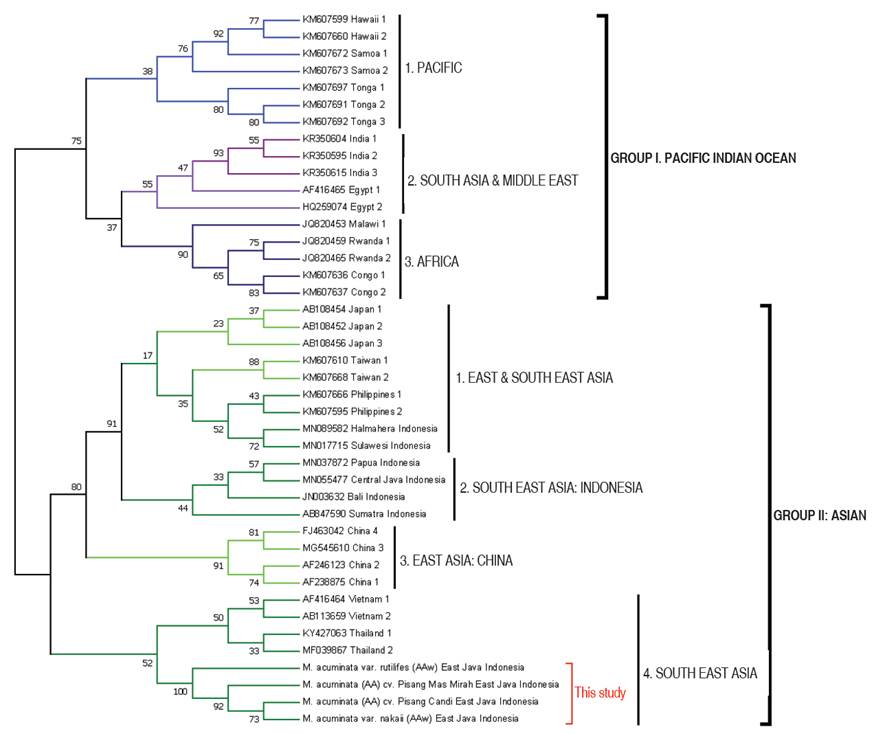
Figure 2 Neighbor-Joining phylogenetic tree of BBTV DNA-R isolates (bootstrap consensus with 50% cut-off).
Specifically, BBTV DNA-R isolates from Indonesia were found separated in three different subgroups based on the source of the materials. Isolates from Sulawesi and Halmahera Islands were clustered in sub-group 1 with the Philippines, Taiwan and Japan. It is thought to be due to its close geographical position. Phylogenetic analysis based on DNA-S and DNA-C analysis also showed that isolates from the islands of Sulawesi and Halmahera were closely related to isolates from Philippines (Rahayuniati et al. 2021b). Furthermore, isolates from Sumatera, Central Java, Bali, and Papua were separated as sub-group 2 (Indonesia only). Meanwhile, isolates from East Java (this study) were nested in sub-group 4 closely related with Thailand and Vietnam isolates (Figure 2). Hence, BBTV DNA-R from East Java were found to be unique and has different route of evolutionary history from the other isolates from Indonesia. Their evolutionary history were presumably came from China mainland via Malay Peninsula route; while the other isolates probably came from the East Asia islands route.
CONCLUSIONS
BBTV DNA-R components were detected in all isolates examined, both symptomatic and asymptomatic. Asymptomatic wild bananas showed shorter length and more variable sequences than symptomatic banana cultivars, which presumably to be related to the resistance mechanism of wild bananas to BBTV. Phylogenetic analysis delineates into two groups i.e. Pacific Indian Ocean group/PIO and Asian group. BBTV DNA-R from East Java were clustered in Asian group. It was found to be unique and has different route of evolutionary history from the other isolates from Indonesia, presumably came from China mainland via Malay Peninsula route. Further researches on the resistance mechanism to BBTV in wild bananas are suggested.