Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales
Print version ISSN 0370-3908
Rev. acad. colomb. cienc. exact. fis. nat. vol.40 no.154 Bogotá Jan./Mar. 2016
https://doi.org/10.18257/raccefyn.277
Doi: http://dx.doi.org/10.18257/raccefyn.277
1 Laboratorio de Micología y Fitopatología, Universidad de Los Andes, Bogotá, Colombia*Corresponding authors:Silvia Restrepo, srestrep@uniandes.edu.co
2 Facultad de Ciencias Naturales y Matemáticas, Universidad del Rosario, Bogotá, Colombia
3 Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, USA
4 Laboratório Nacional de Ciência e Tecnologia do Bioetanol (CTBE), Centro Nacional de Pesquisa em Energia e Materiais (CNPEM), Campinas, São Paulo, Brasil*Corresponding authors: Diego Mauricio Riaño-Pachón, diego.riano@bioetanol.org.br
Received: September 3, 2015. Accepted: March 10, 2016
Abstract
The lack of organisation of monophyletic lineages in the phylogeny and taxonomy of the group formerly known as protists has precluded the understanding of the group's evolutionary history and trait comparison among members of the group. We used a phylogenomic approach to establish phylogenetic hypotheses of this group of organisms. We used an automatic orthologous clustering (OrthoMCL)-based strategy to recover 72 clusters of orthologues from 73 eukaryotic species. A maximum likelihood tree was inferred from the supermatrix. Overall, we obtained consistent inferences with previous published ones, but some unexpected phylogenetic relationships were poorly supported. Despite the large quantity of genes from the Opisthokonta groups, this clade was recovered as polyphyletic. We failed to recover a monophyletic Excavata group, most likely because of long- branch attraction artefacts. A second dataset was constructed after removing the fast-evolving/saturated sites, and a Shimodaira-Hasegawa test was performed to verify whether our data allowed us to reject relationships in previous hypotheses. The results of these tests suggested that the competing tree topologies were not significantly better than our recovered topologies. Novel relationships were shown inside the Opisthokonta, for two species, Thecamonas trahens and Capsaspora owczarzaki. Additionally, some controversial phylogenetic positions among several eukaryotic groups were found. We discuss the relative positions of the Alveolata and Stramenopila groups, the latter being of special interest in our research group.
Key words: Phylogenomics, Markovian Ortholog Clustering, Opisthokonta, Stramenopila, Alveolata.
Resumen
La falta de organización en linajes discretos en la filogenia y la taxonomía del grupo anteriormente llamado protistas ha retrasado la comprensión de la historia evolutiva del grupo y la comparación de rasgos entre los miembros del mismo. En este estudio usamos una aproximación filogenómica para plantear hipótesis filogenéticas del grupo mencionado. Usamos una estrategia basada en el agrupamiento automático de ortólogos (OrthoMCL) para recuperar 72 grupos de ortólogos de 73 especies. Un árbol obtenido con el método de maxima verosimilitud fue estimado a partir de una supermatriz de datos. De manera general obtuvimos inferencias filogenéticas consistentes con publicaciones previas pero se observaron algunos patrones de ramificación inesperados con valores bajos de soporte. A pesar de la gran cantidad de genes de los grupos Opisthokonta, este clado aparece polifilético. No pudimos demostrar la monofilia de Excavata, muy probablemente debido a artefactos de atracción de ramas largas. Un segundo conjunto de datos fue construido luego de eliminar los sitios de rápida evolución/saturados. El test de Shimodaira-Hasegawa se calculó con el fin de verificar si nuestros datos e inferencias filogenéticas controvertían patrones de ramificación reportados previamente. Los resultados de los tests sugieren que las topologías propuestas en estudios previos no son significativamente mejores que las topologías propuestas en este estudio. Nuevas relaciones fueron encontradas dentro de los Opisthokonta, para dos especies, Thecamonas trahens y Capsaspora owczarzaki. Adicionalmente, algunas posiciones filogenéticas controversiales se encontraron para varios grupos eucariotas con nuestra aproximación filogenómica. En el estudio se discuten las relaciones de los grupos Alveolata y Stramenopila, siendo este ultimo grupo de especial interés para nuestro grupo de investigación.
Palabras clave: Filogenómica, Markovian Ortholog Clustering, Opisthokonta, Stramenopila, Alveolata.
Introduction
The group of the organisms formerly known as protists is characterised by the great variety of organisms that are grouped within it. However, the lack of organisation into discrete lineages has been one of the main phylogenetic and taxonomic issues in this group. Protists were eukaryotic organisms with a high diversity in the levels of organisation, comprising unicellular organisms or parenchymatous aggregations. They lack vegetative tissue differentiation (except during reproduction) (Adl, et al., 2007) and are regarded as the group from which multicellular organisms with true tissues differentiated (Adl, et al., 2007; Ruiz-Trillo et al., 2007). Because of the lack of specificity in how "protist" is defined, the taxonomy of the group has been difficult and controversial, raising several issues (Adl, et al., 2005, 2012; Simpson & Roger, 2004).
The classification of the organism previously grouped as protists has always been troublesome because of a number of factors, e.g., random and systematic errors, ambiguous classification criteria and non-flexible systems of classification (Adl, et al., 2007; Adl, et al., 2005; Keeling, et al., 2005; Simpson & Roger, 2004). Historically, the classification of these organisms has sometimes suffered from over-simplification, relying on criteria such as whether the organism was plant-like or animal-like (Keeling, et al., 2005). As a consequence, several species were represented more than once in the classification system (Adl, et al., 2007; Simpson & Roger, 2004). The introduction of a morpho-biochemical approach helped reduce some of these problems. It allowed for the coherent and consistent grouping of most taxa belonging to the group formerly known as protists. The relationships among species inside these groups, for example, within the Alveolata, were consistent with those later reconstructed by molecular methods. The problem, then, was that the evolutionary relationships among the supergoups (as defined in Adl, et al., 2005) still remained unclear because of the lack of a phylogenetic signal in the characters that were used for classification (Adl, et al., 2007; Keeling, et al., 2005).
In the last 30 years, the amount of available molecular data for taxa previously classified as protists has increased, providing useful information from which to infer consistent relationships. Using these resources, scientists have been able to refute former schemes of protist classification and conclude that the rank system used before was inadequate and obsolete. As the amount of data grew, inconsistencies in the classification system became increasingly evident. These inconsistencies, such as the existence of an entire class inside a class (Adl, et al., 2005), suggested that the criteria for grouping and classifying eukaryotic diversity needed to be reconsidered. Thus, a system based on nameless ranked systematics has been proposed, which consists of somewhat abstract categories that are more flexible than ranks (Adl, et al., 2005). Many molecular-based phylogenetic hypothesis including groups formerly classified as protist have been published, but the relationships that have been hypothesized remain controversial due to the inconsistencies between molecular phylogenetic studies (Keeling, et al., 2005).
As mentioned above, difficulties in inferring reliable molecular phylogenies arise from two main sources: i) random error: too little information because of a reductionist approach in the case of single gene-based analyses and long timescales, which gradually deplete phylogenetic signal; ii) systematic error: failure of a phylogenetic method to yield the correct tree because of oversimplified models that are not able to manage the complexity of the evolutionary process of these organisms. When sufficient raw data are provided, it is possible to reliably infer ancient phylogenies (Keeling, et al., 2005). Expressed sequence tags (ESTs) and whole genomes provide a great deal of information and can be used to build a robust phylogenetic matrix (Keeling, et al., 2005).
Phylogenomics, or the use of whole-genome data to infer evolutionary relationships, allows the development of more robust phylogenetic hypotheses because it uses a greater amount of information, overcoming the problem of the lack of phylogenetic signal. Perhaps the strongest advantage of using whole genomes, when compared to the use of ESTs, is that absent markers in the EST dataset are generated because of a lack of data collection and in the genomes they reflect real gains or losses of loci because of evolutionary forces (Leigh, et al., 2011). As databases improve and sequencing techniques become more accessible, the data available for phylogenomics approaches increases greatly, providing new elements for the study of the evolution, genetics and the biology and functionality of increasing number of organisms. We propose a phylogenomic approach to establish phylogenetic relationships among lineages previously classified as protists. To infer the evolutionary relationships among the groups of the organism formerly known as protists and other eukaryotes, such as fungi, animals and plants, we obtained several groups of orthologous genes using a Markov clustering algorithm and then used maximum-likelihood-based phylogenetic reconstruction. Of particular interest for our study group was the position of Chromista Kingdom, in particular the Stramenopila and its relationships within other Eukaryota lineages (Adl, et al., 2005, 2012; Harper & Keeling, 2003; Simpson & Roger, 2004).
Materials and methods
Eukaryotic species considered
We downloaded publicly available deduced proteomes of 77 species to have as many representatives as possible of the major eukaryote groups. We developed a catalogue of the species considered in this study, with their current classification and the source of their proteomes to provide easy access to data. Organisms from the Eukaryota supergoups as defined by Adl, et al. (2005) were included. These supergoups are: Amoebozoa, Opisthokonta, Rhizaria, Archaeplastida, Chromalveolata and Excavata.
The species, their ID and the number of genes for each species were: Eimeria tenella (ETEN, 15), Neospora caninum (NCAN, 18), Toxoplasma gondii (TGON, 20), Cryptosporidium muris (CMUR, 13), Cryptosporidium hominis (CHOM, 12), Cryptosporidium parvum (CPAR, 11), Giardia intestinalis (GLAM, 8), Babesia bovis (BBOV, 18), Theileria annulata (TANN, 14), Theileria parva (TPAR, 14), Plasmodium knowlesi (PKNO, 17), Plasmodium vivax (PVIV, 16), Plasmodium falciparum (PFAL, 16), Plasmodium chabaudi (PCHA, 16), Plasmodium berghei (PBER, 16), Plasmodium yoelii (PYOE, 16), Leishmania braziliensis (LBRA, 16), Leishmania mexicana (LMEX, 16), Leishmania infantum (LINF, 16), Leishmania major (LMAJ, 16), Trypanosoma cruzi (TCRU, 12), Trypanosoma vivax (TVIV, 16), Trypanosoma brucei (TBRU, 15), Trypanosoma congolense (TCON, 14), Selaginella moellendorffii (SOME, 46), Arabidopsis lyrata (ALYR, 44), Sorghum bicolor (SBIC, 47), Coccomyxa sp (CSP, 37), Chlorella vulgaris (CVUL, 37), Micromonas pusilla (MPUS, 40), Ostreococcus lucimarinus (OLUC, 33), Bigelowiella natans (BNAT, 45), Cyanidioschyzon merolae (CMER, 19), Guillardia theta (GTHE, 44), Emiliania huxleyi (EHUX, 30), Aureococcus anophagefferens (AANO, 22), Fragilariopsis cylindrus (FCYL, 17), Phaeodactylum tricornutum (PTRI, 24), Phytophthora capsici (PCAP, 46), Phytophthora ramorum (PRAM, 41), Phytophthora sojae (PSOJ, 46), Naegleria gruberi (NGRU, 41), Dictyostelium purpureum (DPUR, 47), Entamoeba invadens (EINV, 11), Entamoeba dispar (EDIS, 13), Entamoeba histolytica (EHIS, 12), Enterocytozoon bieneusi (EBIE, 2), Nosema ceranae (NCER, 4), Encephalitozoon cuniculi (ECUN, 3), Enterocytozoon hellem (EHEL, 2), Encephalitozoon intestinalis (EINT, 4), Thecamonas trahens (TTRA, 43), Trichomonas vaginalis (TVAG, 13), Allomyces macrogynus (AMAC, 18), Batrachochytrium dendrobatidis (BDEN, 38), Mucor circinelloides (MCIR, 40), Phycomyces blakesleeanus (PBLA, 36), Auricularia delicata (ADEL, 29), Agaricus bisporus (ABIS, 27), Acremonium alcalophylum (AALC, 25), Aspergillus niger (ANIG, 26), Sphaeroforma arctica (SARC, 37), Monosiga brevicolis (MBRE, 43), Salpingoeca roseta (SROS, 50), Capsaspora owczarzaki (COWC, 44), Trichoplax adhaerens (TADH, 55), Nematostella vectensis (NVEC, 55), Daphnia pulex (DPUL, 54), Capitella teleta (CTEL, 58), Lottia gigantea (LGIG, 57), Ciona intestinalis (CINT, 47), Canis familiaris (CFAM, 47), Homo sapiens (HSAP, 45). The species' names are represented by an ID and encoded as following: The first letter corresponds to the first letter of the genus and the three remaining correspond to the three first letters of the specific epithet.
Phylogenomic workflow
Clustering of Orthologous genes. An all-versus-all BlastP search (Altschul, et al., 1997) was performed on all of the protein sequences (cut-off E-value = 10-5) to obtain prior similarity tables as input for the Markov cluster (MCL) algorithm. To construct orthologous groups, we used the OrthoMCL package because it provides a method of grouping orthologous genes across multiple eukaryotic taxa (Li, Stoeckert, and Roos, 2003) and because it has been shown to perform best in terms of the balance of sensitivity and the specificity of orthologous detection (Chen, Mackey, Vermunt, and Roos, 2007). The orthologous detection algorithm was run with three different inflation values (I = 1.2, 1.5, 2.0). Greater inflation values yield clusters with a lesser number of genes (tighter) and a greater number of these clusters (Chen, et al., 2007). From the entire set of orthologous clusters, we kept only those containing unique copies of orthologous genes in each species. Phylogenetic analyses were performed only on clusters derived from the I = 1.5 run because this value yielded the most populated groups in terms of number of species.
Phylogenetic reconstruction. For each cluster of orthologous proteins, we performed multiple sequence alignments using MAFFT, parameters by default (Katoh, Kuma, Toh, and Miyata, 2005) The evolutionary model for every cluster was then determined using ProtTest (Abascal, Zardoya, and Posada, 2005). A supermatrix was built using FASconCAT that included all of the groups of detected orthologues (Kuck & Meusemann, 2010). Finally, phylogenetic inference was performed using the maximum likelihood method implemented in FastTree (Price, Dehal, and Arkin, 2009) using the only evolutionary model available in this package (WAG). We rooted the tree using the midpoint method because of the unavailability of a defined outgroup in our dataset (Hess & De Moraes Russo, 2007). We performed 1000 bootstrap replicates for statistical support, and the bootstrap support values are shown in a maximum scale of 1.
Phylogenetic trees and multiple sequence alignments are available at http://bce.bioetanol.cnpem.br/protistphylogenomics.
Taxonomical assessment of the monophyletic lineages in the group formerly known as protists
After revising the evolutionary lineages found in the phylogenetic reconstruction, we aimed to reconstruct the taxonomical ranks between the group of organisms formerly known as protists. We used the taxonomical ranks proposed by Adl, et al. (2005, 2012) and followed their guidelines in the organisation of those ranks. In the case of discrepancies between our reconstruction and the taxonomy proposed, we used the information in our tree to define new taxonomical ranks in which a monophyletic lineage with a clearly different phylogenetic relationship as previously published, should be considered a novel taxonomical category.
The procedure was initially performed with 73 species including the Microsporidia but excluding some oomycetes and green algae. A second procedure (77 species) was performed including the previously omitted species but excluding the Microsporidia and three species in which the long-branch attraction artefact was observed. A third procedure was performed excluding several species from taxa that do not belong to Stramenopila or the Alveolata while including a few representatives of each major supergroup to retain eukaryotic diversity in the dataset.
Functional identification of orthologous groups
To identify the functions of the genes contained in the clusters of orthologues, the PANTHERDB (Protein Analysis Through Evolutionary Relationships) database was used (Mi, et al., 2005). A BlastP (cut-off E-value = 10-5) was performed on our dataset against the PANTHER database to obtain a filtered table of possible hits. Then, we compared the PANTHER database with the BlastP results using the pantherScore tool, also obtained from the PANTHERDB website.
Hypothesis testing and removal of fast-evolving/saturated sites
The test proposed by Shimodaira and Hasegawa (1999) (SH), implemented in the RAxML 7.2.8 package (A. Stamatakis, Heidelberg, Germany), was used to compare our results with three different topologies resulting from two prior hypotheses: i) The Stramenopila is the sister group to the Alveolata, and ii) the genera Giardia, Naegleria and Trichomonas branch within the Excavata. Additionally, fast- evolving and saturated sites that might have been adding noise to our dataset were removed by using the Gblocks package (Castresana, 2000), adjusting the maximum number of non-conserved amino acid positions to 70 and the minimum block length to 10. This adjustment allowed for the conservation of all of the previous alignments' gaps. This latter tree was also compared against the same three topologies produced by the hypotheses mentioned above.
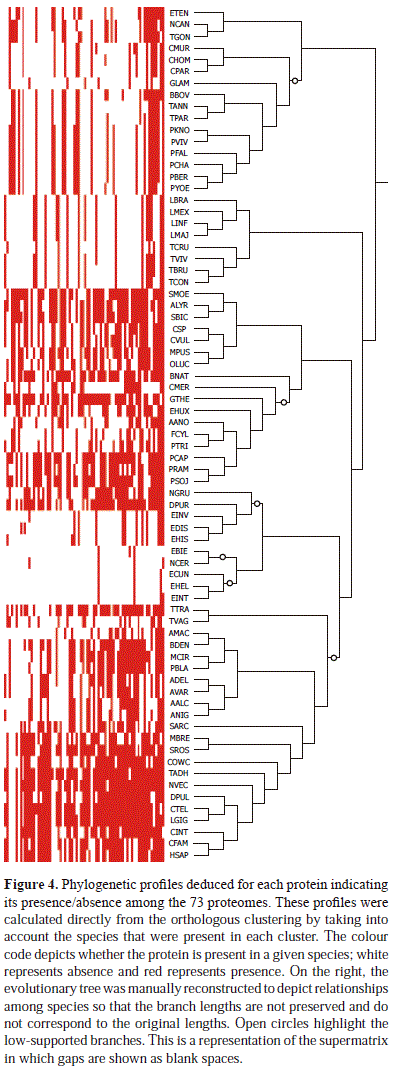
Construction of phylogenetic profiles
Phylogenetic profiles were constructed for each protein deduced from the Markovian clustering algorithm. Clusters of orthologous genes containing the species in which they are present were directly used to address the occurrence of a protein in certain species' proteome. "The phylogenetic tree and underlying alignment were deposited in TreeBase under the accession number HYPERLINK
http://purl.org/phylo/treebase/phylows/study/TB2:S18195"TB2:S18195".Results
The orthologous genes found are spread across the 73 species, and not all of the species are equally represented in terms of the number of genes per species (see Materials and Methods). For example, Microsporidia clade had the fewest genes by contrast to the metazoans, which have the highest number of recovered genes per species.
Unexpected phylogenetic relationships inside the Opisthokonta
After obtaining the tree with bootstrap (BS) support for each clade (Figure 1), we mapped each species to its corresponding supergroup according to the classifications made by Adl, et al. (2005). Representatives of the 6 supergroups were obtained from our 73-species data set. Monophyletic groups were highlighted with blue lines, and non-monophyletic groups were highlighted with red lines. The BS values in red (< 0.7) were considered to be too weak to support the consistency of a clade. Three species are highlighted with a red star. These species belong to the supergroup Excavata according to Adl, et al. (2005), but our tree depicted them as being related to other eukaryotic groups far from the Excavata. They were not taken into account for defining groups whether they are included with highly supported branches or not.
As observed in figures 1 and 2, the clade that contains the metazoans, Capsaspora owczarzaki, the choanozoans and the mesomycetozoans (MCCM clade) was recovered as monophyletic, and the relationships inside it were consistently supported. The closest group to the MCCM clade was the clade comprising all the fungi except the Microsporidia (FWM clade). The branch leading to these two groups had a high BS value indicating strong support of their relationship. Finally, the sister clade to to these two clades was a clade containing two species, Trichomonas vaginalis, which belongs to the Excavata and Thecamonas trahens, the only representative of the Apusozoa in our analysis. This clade (Thecamonas threatens + Trichomonas vaginalis) and the MCCM + FWM clade were not related, but the BS support value (0.667) was near the acceptable 0.7 threshold. The clade that comprises the Amoebozoa and the Microsporidia (AM) was the sister group to the previously mentioned TT + MCCM + FWM clade. Relationships between the Amoebozoa and the Microsporidia were also poorly supported, as were several relationships inside the Microsporidia. The Opisthokonta supergroup was composed of the clades TT+ MCCM + FWM plus the Microsporidia.
Stramenopiles are the closest relatives of Viridiplantae and other algae
Figures 1 and 2 show that the Stramenopila clade appears well-resolved, with strong support on all of its branches, and it is represented by four genera: Phytophthora, Phaeodactylum, Fragilariopsis and Aureococcus. This clade was most closely related to Emiliania (Haptophyta, 0.999 BS), and the Stramenopila + Haptophyta clade was most closely related to Guillardia (Cryptophyta, 0.978 BS), forming the Stramenopila + Haptophyta + Cryptophyta (SHC) clade. The Rhodophyta clade, represented by Cyanidioschyzon merolae, appeared as the sister group of the SHC clade but with poor support (0.561). Additionally, this entire clade was related to the Rhizaria supergroup, represented only by the cercozoan Bigelowiella natans, with a relatively high support value (0.798 BS). The group that comprises the Stramenopila, the Haptophyta, the Cryptophyta, the Rhodophyta and the Cercozoa (Rhizaria) was the sister group of the Viridiplantae clade (0.981 BS), and the monophyly of the Viridiplantae clade, as well as the relationships inside it, were all strongly supported (BS > 0.991). Our data revealed that the Alveolata and the species belonging to the Stramenopila group do not share a recent common ancestor. In fact, according to our data, the Alveolata is the earliest diverging eukaryotic lineage.
Excavata and the earliest diverging lineage, the Alveolata, a supergroup that appears in a controversial position
The Excavata, represented by the genera Leishmania and Trypanosoma, was the sister group of the clade comprising the groups of the Rhizaria and the Archaeplastida and the smaller groups of the Cryptophyta, the Haptophyta and the Stramenopila. This relationship, and the relationships inside the Excavata, was strongly supported (BS > 0.999). The Excavata in the tree depicted in figure 1 (and figure 2) is paraphyletic because it does not include the three species denoted with a red star that other studies have placed in the Excavata (Adl, et al., 2005).
Finally, the Alveolata clade appeared as the first diverging lineage. This clade comprised the genera Eimeria, Neospora, Toxoplasma, Cryptosporidium, Babesia, Theileria and Plasmodium. It also included Giardia as a member, attached as a sister group to the Plasmodium + Theileria + Babesia clade, with relatively high support (0.742 BS), but Giardia was previously reported to belong to the Excavata cluster (Adl et al., 2005). All relationships inside this clade were well-supported (BS > 0.742), except the one between the clade containing the genus Cryptosporidium and the clade that comprises the genera Giardia, Babesia, Theileria and Plasmodium (0.509 BS).
Fast-evolving and saturated sites removed
The phylogenetic tree in figure 3 was built by removing fast- evolving and saturated sites. It depicts similar relationships to the first tree inside the major groups, however BS supports suffered slight changes, and one can find more poorly supported clades (< 0.7) in this new tree. Perhaps the major difference between the two tree topologies is that the earliest diverging group changes from one group in one tree to another group in the other tree. Whereas in the first tree (Figures 1 and 2) the Alveolata clade is the most external group, the second tree (figure 3) places the Microsporidia + Trichomonas + Giardia as the external group, which indicates that the alignment trimming placed several fast-evolving lineages together. Another major change found in this tree is that the genus Thecamonas was placed outside the Opisthokonta and was grouped as a sister group to the Alveolata. Finally, the genus Capsaspora appears as a sister group to the genus Sphaeroforma, forming a monophyletic clade (Choanomonada).
Long-branch artefacts and the Chromalveolata hypothesis
We were particularly interested in: 1) the genera Giardia, Trichomonas and Naegleria because they have not been grouped near their Excavata relatives in any of the trees, and because they are known to produce long-branch attraction artefacts; 2) the Stramenopila and the Alveolata groups because their relationship was well-supported in previous reports (Hampl, et al., 2009) and our reconstruction failed to group them; 3) the Microsporidia clade because of their low genomic representation in our study (4 genes in a species as a maximum), and because although several studies place them as a basal fungi group, our analysis failed to group them as expected, most likely because the few genes that we used may be in regions of fast evolution, as the branches of these species were relatively long. This finding gave rise to two a priori hypotheses to be compared with our results: one that includes the fast-evolving taxa (Trichomonas, Giardia and Naegleria belong to the Excavata, Microsporidia occurs within the fungi), and another that states that the Stramenopila and the Alveolata are more closely related to one another than to any other clade in the dataset. We decided to test whether the topologies associated with these hypotheses were significantly different from our resulting topologies, both in independent topologies and a combined topology of both hypotheses. The Shimodaira-Hasegawa test on the raw and Gblocks trimmed alignments and their corresponding tree topologies showed that none of these hypothetic topologies was significantly better than those obtained with our data set.
Functional identification of orthologous groups
As mentioned above, the number of orthologous genes were not the same in all species examined. This is also displayed as a phylogenetic profile (Figure 4) in which the presence/absence of a given gene in a determined species is coded by red/white.
Twenty-five protein families' biological functions were successfully identified by comparing our data against the PANTHER database using hidden Markov model- based tools. Proteins such as MYB transcription factors (PTHR13856:SF31), DNA polymerases (PTHR10133), elongation factors (PTHR23115:SF66) and DNA repair proteins (PTHR10799, PTHR22850:SF13), cell mem- brane proteins (PTHR10795), G proteins, Transferases (PTHR11135, PTHR32119:SF2, PTHR21329), Hydrolases (PTHR11820:SF77), DNA helicase (PTHR10799:SF213), Heat Shock proteins (PTHR11528), Transducin Beta-like protein (PTHR19854:SF15), a signal recognition particle 9kd protein (PTHR12834), a potassium voltage-gated channel protein (PTHR10217:SF376), SNRNA-activating protein complex subunit 3 (PTHR13421), MUTS homolog 4, MSH4 (PTHR11361:SF36), Adapter-related protein complex, beta subunit (PTHR11134), Bartet-Biedl syndrome proteins (PTHR23083:SF389), kinase (PTHR12400), Tumour necrosis factor type 1 receptor associated protein (PTHR11528) and several other hypothetical and putative proteins (PTHR12895, PTHR15830:SF5, PTHR22957, PTHR15840:SF4) were found in our dataset.
Further analyses modifying the number of taxa also contribute evidence to refute the Chromalveolata hypothesis
Two additional trees were inferred from datasets that were built by varying the number of species included. The first included 77 species, including more species from the Archaeplastida and the Stramenopila than the previous dataset. Additionally, the microsporidian species, and those species previously reported to cause LBA were removed. The second dataset had only 53 species, retaining all of the taxa from the Stramenopila and the Alveolata but discarding several taxa from other supergroups, leaving only a few representatives of each group. The first of these datasets was built to improve taxon sampling, and the second one was built to improve gene clustering because it seems to be a compromise between species' divergence and performance of the MCL. With these new trees, we expected to obtain a more robust view of the Stramenopila + Alveolata hypothesis (the Chromalveolata hypothesis).
The first tree (Figure 5) yielded a topology in which the Alveolata appears as the earliest diverging lineage, and the Stramenopila (without Guillardia theta) + Cyanidioschyzon merolae appears as the closest relative to the Excavata + Entamoeba clade, but this relationship was poorly supported. This clade appeared as the sister group of the Rhizaria. It is also observed that Viridiplantae + Guillardia theta + Thecamonas trahens is most closely related to Dictyostelium + Opisthokonta. The second tree (Figure 6) yielded a topology that resembles that of figure 1. Again, the Alveolata, as in almost all of the previous trees, appeared as the earliest diverging lineage. The rest of the topology is similar to the figure 1 tree, except that the Rhizaria lineage diverges earlier, and Entamoeba dispar does not group with the genus Dictyostelium but appears on an early diverging branch.
Discussion
This study implements large datasets in complex computational algorithms for the reconstruction of the evolutionary history of the group formerly known as protists. Several studies have used different genes to reconstruct the phylogeny of this group ofeukaryots (Burki, et al. 2009; Cavalier-Smith & Chao, 2010), but here we implemented a Markov clustering method to optimise and debug the dataset of genes to develop a robust phylogeny. The use of this complex computational method and the use of a number of eukaryotic complete proteomes that are publicly available allow this research to provide a deeper insight into the phylogeny and taxonomy of the organisms classified in this diverse and poorly understood group.
With our novel approach, the phylogenetic relationships of several species were resolved. The Opisthokonta was recovered as a polyphyletic group, and inside it, Capsaspora sp. was found to be the lineage closest to the metazoans. This is a novel phylogenetic hypothesis for metazoans and Capsaspora sp., not previously observed in traditional taxonomies. The lineage represented by Thecamonas sp., previously ranked as of uncertain origin, appears as the closest relative to the Opisthokonta. Our analysis also places the stramenopiles, haptophytes and cryptophytes inside a monophyletic lineage (Cavalier-Smith & Chao, 2006), going against the Chromalveolata hypothesis, and places the rhodophytes, rhizarians and Viridiplantae lineages as their closest relatives instead of the Alveolata group, which was previously reported to be the closest group to the Stramenopila (Adl, et al., 2005; Burki, et al., 2009). The only representatives of the Excavata that were grouped as a monophyletic clade were the Leishmania and Trypanosoma species (Kinetoplastids) (Adl, et al., 2005), whereas other Excavata members are dispersed throughout other clades in the tree.
Our analysis did not support the monophyly of almost any of the so-called traditional supergroups. The dashed lines in our tree (Figure 1) trace and indicate the supergroups to which each clade belongs according to the previously mentioned classification system. While we are aware that some of those placements may have occurred because of the limitations of our procedure, most of our results were well-supported and consistent, solving previously unclear relationships, and reaffirming formerly published relationships.
Apparent Opisthokont polyphyly and the position of the Microsporidia
The five microsporidian species are the only ones that group outside the Opisthokont clade, but they were expected to share a common ancestor with the fungi (Corradi & Keeling, 2009). We found that these species have the least number of representatives in our dataset because they had, at most, four orthologous genes that were recovered and included in the supermatrix. This can also be observed in the phylogenetic profile (Figure 4), in which the differences between the phylogenetic profile of microsporidian species and other fungi are remarkable. The microsporidia are obligate intracellular parasites of other eukaryote species, most frequently animals. They lack several cellular components, such as mitochondria, the Golgi apparatus, and centrioles, and this can be correlated to a reduced genome that might be a result of high levels of specialisation. In fact, it is reported that they have approximately 2000 genes that include several fast-evolving and divergent sequences that might be causing long branch attraction artefacts in some analyses (Brinkmann, van der Giezen, Zhou, Poncelin de Raucourt, and Philippe, 2005; Corradi & Keeling, 2009). This could contribute to the high levels of noise in our dataset, producing misleading results. Moreover, three of the four amoebozoa are categorised as low-sampled species, which can add the effect of long-branch attraction to the few sequences obtained from the microsporidians. Despite the apparent polyphyly of the Opisthokonta, caused by LBA, as mentioned previously, our tree reveals and confirms the Amoebozoa as the closest relatives of the Opisthokonta, as found in previous analyses (Hampl, et al., 2009; Ruiz-Trillo, et al., 2007).
The positions of Capsaspora owczarzaki and Thecamonas trahens
Capsaspora was found to be included in 3 groups also including several metazoan species and in which the choanozoans (Monosiga and Salpingoeca) were not included. Capsaspora was reported to share more transcription factors with metazoans than with the genus Monosiga (Sebe-Pedros, de Mendoza, Lang, Degnan, and Ruiz-Trillo, 2011).
The genus Thecamonas was previously reported to belong to a group of uncertain origin (Apusomonadida) (Adl, et al., 2005). Our analysis placed it as a sister group of the Opisthokonta (excluding the Microsporidia), with a support value (0.667 BS), slightly below the acceptable 0.7 threshold, indicating that it shares a common origin with the Opisthokonta. This result is consistent with a previously reported classification of this organism and reinforces this classification because it had been only built based on a single gene (Cavalier-Smith & Chao, 2010).
Controversy regarding the monophyly of the Chromalveolata
The Stramenopila, the Cryptophyta, the Haptophyta and the Alveolata were previously reported to share a common ancestor and to constitute the Chromalveolata clade (Adl, et al., 2005; Burki, et al., 2009). However, our analysis contends such monophyly and places the Stramenopila, the Haptophyta and the Cryptophyta together. These three form the former group Chromista (Cavalier-Smith & Chao, 2006), and the immediate relatives of the Stramenopiles in our tree are the red algae Cyanidioschyzon merolae, the rhizarians and then the Viridiplantae. These results coincide with other phylogenomic analyses performed by different methods of evaluation of orthologous genes (Ocana & Davila, 2011). The genes recovered with the Markovian clustering for these lineages show that the Stramenopila and the Alveolata do not share a recent common ancestor that allows them to constitute a monophyletic group. We suggest further revision of the phylogenetic relatoinships of the Stramenopila and the Alveolata, along with the addition of several Archaeplastica species.
The divergence of the Rhodophyta from the Viridiplantae, and the failure of the reconstruction of a monophyletic Archaeplastida, can also be attributed to the low number of genes in the genus Cyanidioschyzon and a possible retention of red algae nuclear genes by the cryptophytes (Brinkmann, et al., 2005).
When evaluating the additional datasets that were generated and the trees associated with them (Figures 5 and 6), topologies consistent with those of the previous trees were found, suggesting that the Stramenopila and the Alveolata do not constitute a monophyletic group. Our trees also show that the Stramenopila and the Alveolata do not constitute a monophyletic group, a result that is opposed in several previously reported phylogenies (Baurain, et al., 2010; Burki, et al., 2009; Hampl, et al., 2009; Hess & De Moraes Russo, 2007; Parfrey, et al., 2010). This makes our results different from other studies and do not support the Chromalveolata hypothesis (Bodyl, Stiller and Mackiewicz, 2009; Burki, et al., 2009). The phylogenetic relationships that were found here link the stramenopiles with plants and green algae (Viridiplantae) more than with any other lineage, and they place the Alveolata as the earliest diverging lineage. Although we cannot reject the Chromalveolata hypothesis based on the result of the Shimodaira-Hasegawa test alone, the strong support obtained for most of the branches in our trees, and the consistent position of the Alveolata, even when the dataset was changed, allow us to state that the Stramenopila and the Alveolata are not sister groups. Figure 4 shows that the phylogenetic profiles of the Stramenopila and the Alveolata groups are quite dissimilar and that the Alveolata profile resembles the Excavata profile more than it resembles the Stramenopila profile. Additionally, the Stramenopila profile is similar to the Viridiplantae profile. The hypothesis that suggests that the Stramenopila and the Alveolata share a red algal common ancestor that originated by a single endosymbiotic event (Simpson & Roger, 2004) is not supported.
Giardia, Trichomonas and Naelgeria
The Excavata also appear in Figure 4 as a group whose representatives are considered to be scarcely represented. Most of the Excavata organisms (6 species, 3 from the genus Leishmania and 3 belonging to the genus Trypanosoma) belong to a subgroup of the Euglenozoa called the Kinetoplastea, which are characterised, so far, by the presence of a mass of DNA associated with their flagellar bases (kinetoplast) (Adl, et al., 2005). Giardia lamblia and Trichomonas vaginalis are reported to be species with fast-evolving sites in their genomes (Hampl, et al., 2009) that can contribute error to datasets in which they are included because fast-evolving sites are strongly associated with long branch attraction artefacts (Philippe, et al., 2000). Although no information relating species of Naegleria with these phenomena were found, we cannot discard the possibility that this phenomenon occurs in our dataset. Another contributing factor to the misplacement of Naegleria gruberi could be that the number of genes present in this species in our dataset (41) differs greatly from the number of genes recovered from the other members of the Excavata (16 or less). At this point, we cannot dismiss that these genera belong to the Excavata or that the Microsporidia belongs to the fungi. The latter confirms that the long-branch artefact might have affected the relationships of these and several other species.
Removing fast-evolving/saturated sites
The trimmed dataset produced some similar topologies and overall lower statistical support compared to our original dataset. This dataset was also compared to the three hypothesis-based topologies via SH test, yielding identical results as the comparison of the original one. This indicates that, in this case, trimming the gene alignments and concatenating them afterward produces additional gaps. Filling blank regions that are caused by the performance of the clustering algorithm did not result in a gain of phylogenetic signal. To yield an improved dataset so that the trimmed dataset would contain less additional gaps, it might be beneficial to manually complete the gene stock for several poorly represented species, taking into account several other sources of orthologous eukaryotic genes, such as ESTs, and then performing the removal of these noise-adding sites.
Initially, two eukaryotic supertrees were reconstructed based on an automatic orthologous clustering approach; one with no further supermatrix modifications, and one with the fast-evolving and saturated sites removed. It was shown that Capsaspora owczarzaki is the closest relative of the Metazoa clade. It was previously suggested that the origin of Thecamonas trahens was uncertain (Adl, et al., 2005). Here we confirm that it shares a common ancestor with the Opisthokonta and suggest that they are placed in the same supergroup; these two results have not been previously reported. The Stramenopila and the Alveolata were not confirmed to be sister lineages as in previous analyses; thus, further revision of these clades' phylogenetic relationships are recommended. The Excavata was found to be monophyletic except for three species, two of them previously reported as generators of a long-branch attraction artefact. Despite the efforts of removing the saturated/ fast-evolving blocks, their correct placement was not achieved. This is attributed to the difference in the amount of genes per species.
Automatic orthologue clustering was shown to be a remarkably efficient approach to recover homologous proteins and to build consistent datasets that yield enough phylogenetic information to unravel several unclear relation-ships among supergroups of eukaryotic organisms. Problems found with this approach are attributed to the choice of species because their natural histories may have resulted in genome size reduction. This reduction affects the quality of the gene grouping and can be sorted using additional data, such as the recovery of ESTs and the completion of the genomic databases, which can provide valuable information for studying orthologous genes and the functionality and evolution of this intricate group of organisms.
Acknowledgements
We thank Dr. Andrew J. Crawford, Francisco Buitrago and David Urbina for their assistance and suggestions. We also thank the Department of Biological Sciences of the Universidad de Los Andes, Bogotá for providing technical resources to make this work possible.
Conflict of interests
The authors declare no having any conflict of interest in publishing this article.
Bibliography
Abascal, F., Zardoya, R., and Posada, D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics, 21: 2104-2105. [ Links ]
Adl, S.M., Simpson, A.G., Farmer, M.A., Andersen, R.A., Anderson, O.R., Barta, J.R., et al. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol., 52: 399-451. [ Links ]
Adl, S.M., Leander, B.S., Simpson, A.G., Archibald, J.M., Anderson, O.R., Bass, D., et al. (2007). Diversity, nomen- clature, and taxonomy of protists. Syst Biol. 56: 684-689. [ Links ]
Adl, S.M., Simpson, A.G., Lane, C.E., Lukes, J., Bass, D., Bowser, S.S., et al. (2012). The Revised Classification of Eukaryotes. J. Eukaryot. Microbiol., 59: 429-493 [ Links ]
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI- BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25: 3389-3402. [ Links ]
Baurain, D., Brinkmann, H., Petersen, J., Rodríguez-Ezpeleta, N., Stechmann, A., Demoulin, V., et al. (2010). Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol., 27: 1698-1709. [ Links ]
Bodyl,A., Stiller, J.W., and Mackiewicz, P. (2009). Chromalveolate plastids: direct descent or multiple endosymbioses? Trends Ecol Evol., 24: 119-121; author reply 121-112. [ Links ]
Brinkmann, H., van der Giezen, M., Zhou, Y., Poncelin de Raucourt, G., Philippe, H. (2005). An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst Biol. 54: 743-757. [ Links ]
Burki, F., Inagaki, Y., Bråte, J., Archibald, J.M., Keeling, P.J., Cavalier-Smith, T., et al. (2009). Large-scale phylogenomic analyses reveal that two enigmatic protest lineages, telonemia and centroheliozoa, are related to photo-synthetic hromalveolates. Genome Biol Evol., 1: 231-238. [ Links ]
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol., 17: 540-552. [ Links ]
Cavalier-Smith, T., & Chao, E.E. (2006). Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J Mol Evol, 62: 388-420. [ Links ]
Cavalier-Smith, T., & Chao, E.E. (2010). Phylogeny and evolution of apusomonadida (protozoa: apusozoa): new genera and species. Protist, 161: 549-576. [ Links ]
Chen, F., Mackey, A.J., Vermunt, J.K., and Roos, D.S. (2007). Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE, 2: e383. [ Links ]
Corradi, N., & Keeling, P.J. (2009). Microsporidia: a journey through radical taxonomical revisions. Fungal Biol Rev., 23: 1-8. [ Links ]
Hampl, V., Hug, L., Leigh, J.W., Dacks, J.B., Lang, B.F., Simpson, A.G., et al. (2009). Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic "supergroups". Proc Natl Acad Sci U S A., 106: 3859-3864. [ Links ]
Harper JT., & Keeling, P.J. (2003). Nucleus-encoded, plastid-targeted glyceraldehyde-3-phosphate dehydrogenase (GAPDH) indicates a single origin for chromalveolate plastids. Mol Biol Evol., 20: 1730-1735. [ Links ]
Hess, P.N., & De Moraes Russo, C.A. (2007). An empirical test of the midpoint rooting method. Biol J Linn Soc Lond., 92: 669-674. [ Links ]
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res., 33: 511-518. [ Links ]
Keeling, P.J., Burger, G., Durnford, D.G., Lang, B.F., Lee, R.W., Pearlman, R.E., et al. (2005). The tree of eukaryotes. Trends Ecol Evol. 20: 670-676. [ Links ]
Kuck, P., & Meusemann, K. (2010). FASconCAT: Convenient handling of data matrices. Mol Phylogenet Evol., 56: 1115- 1118. [ Links ]
Leigh, J.W., Lapointe, F.J., Lopez, P., and Bapteste, E. (2011). Evaluating phylogenetic congruence in the post-genomic era. Genome Biol Evol., 3: 571-587. [ Links ]
Li, L., Stoeckert, C.J., Jr., Roos, D.S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res., 13: 2178-2189. [ Links ]
Mi, H., Lazareva-Ulitsky, B., Loo, R., Kejariwal, A., Vandergriff, J., Rabkin, S., et al. (2005). The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res., 33: D284-288. [ Links ]
Nozaki, H., Maruyama, S., Matsuzaki, M., Nakada, T., Kato, S., and Misawa, K. (2009). Phylogenetic positions of Glaucophyta, green plants (Archaeplastida) and Haptophyta (Chromalveolata) as deduced from slowly evolving nuclear genes. Mol Phylogenet Evol., 53: 872-880. [ Links ]
Ocana, K.A., & Davila, A.M. (2011). Phylogenomics-based reconstruction of protozoan species tree. Evol Bioinform Online, 7: 107-121. [ Links ]
Parfrey, L.W., Grant, J., Tekle, Y.I., Lasek-Nesselquist, E., Morrison, H.G., Sogin, M.L., et al. (2010). Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol., 59: 518-533. [ Links ]
Philippe, H., Lopez, P., Brinkmann, H., Budin, K., Germot, A., Laurent, J., et al. (2000). Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc Biol Sci., 267: 1213-1221. [ Links ]
Price, M.N., Dehal, P.S., and Arkin, A.P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol., 26: 1641-1650. [ Links ]
Ruiz-Trillo, I., Burger, G., Holland, P.W., King, N., Lang, B.F., Roger, A.J., et al. (2007). The origins of multicellularity: a multi-taxon genome initiative. Trends Genet., 23: 113-118. [ Links ]
Sebe-Pedros, A., de Mendoza, A., Lang, B.F., Degnan, B.M., Ruiz-Trillo, I. (2011). Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol., 28: 1241-1254. [ Links ]
Shimodaira, H., Hasegawa, M. (1999). Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol., 16: 1114. [ Links ]
Simpson, A.G.B., & Roger, A.J. (2004). The real 'kingdoms' of eukaryotes. Cambridge, MA: Cell Press. [ Links ]













