Introduction
Antibiotics have undoubtedly improved health and life expectancy and have revolutionized modern medicine. These medications allow the control of previously untreatable bacterial infections and have increased the therapeutic success of high-risk medical interventions such as surgeries and chemotherapies. Despite their great success, we are currently facing a crisis due to the emergence of antibiotic-resistant microorganisms that represent a risk to global human health and, thus, require immediate measures to prevent therapeutic failure and ensure a continuous and effective treatment of bacterial infections.
The worldwide menace posed by resistant pathogens has led to the definition of strategies for the comprehensive management of antibiotic resistance (World Health Organization-WHO, 2016). One of the priorities is centered on the control of healthcare-associated infections, particularly when multidrug-resistant (MDR) microorganisms are involved (Ventola, 2015). Some of the pathogens that are critical in hospitals include methicillin-resistant Staphylococcus aureus (MRSA), Enterococcus species resistant to vancomycin, and Enterobacteria such as Klebsiella pneumoniae (Kpn), Pseudomonas aeruginosa, and Acinetobacter. Besides limiting therapeutic options, antimicrobial resistance also represents an economic burden to health systems in general as it increases the costs associated with treatment both at the global and local scales (Barrero, 2014; Cosgrove, 2006; Ortiz-Mayorga, 2019). In Colombia, the prevalence and dissemination of antimicrobial resistance in hospitals has also led to proposals aimed at controlling resistant pathogens using strategies that include early detection, surveillance, and the prudent use of antibiotics (Leal, 2016; Ministerio de Salud, 2018; Rada, 2019).
Although we know today that antibiotic resistance is inevitable (Ventola, 2015), certain conditions can accelerate the selection of resistant microbes. The excessive and inappropriate use of antimicrobials for human health, agriculture, and industry (Carlet, 2012) has increased the amount of these drugs in the environment where they can impose selective pressure on bacteria as they adapt to survive, generating conditions for the selection of resistance. The combination of resistance and virulence factors, cell determinants that allow a microbe to cause disease, can be particularly lethal, especially when dealing with MDR bacteria (Rodríguez-Rojas, 2013). Besides the inadequate use of antimicrobials, today we face a scarcity of promising new drugs and lead compounds in the development pipeline (Pendleton, 2013). The high costs of discovery, coupled with the low economic returns and strict regulatory requirements, have reduced interest in the development of novel antimicrobial compounds (Ventola, 2015).
Microorganisms and their antibiotic resistance genes (ARG) are ubiquitous and diverse (Allen, 2010). Many studies on ARGs present in natural environments have been done using metagenomics in which analysis of genes and genomes replaces the culturing of microorganisms. This approximation can provide information on both the taxonomic profiles and the metabolic repertoire of the microbial communities studied. Based on these analyses, we know that ARGs are present in intact and relatively pristine ecosystems such as the ancestral permafrost (D'Costa, 2011), Alaskan soils and cave communities (Allen, 2009; Bhullar, 2012), as well as in the human microbiome and in more heavily contaminated sites such as lakes, rivers, and wastewater treatment plants (Marti, 2014; Posada-Perlaza, 2019; Rizzo, 2013).
Antimicrobial resistance is gained either by mutations or by the acquisition of genes via horizontal gene transfer (HGT). Microorganisms have the genomic plasticity and capacity to access genetic material from other microbes and share genes, such as ARGs (Gaze, 2013) . The transfer of genetic information among bacteria is facilitated by mobile genetic elements like plasmids, transposons, and bacteriophages. In fact, the spread of plasmids bearing ARGs has been known to occur in hospital settings for many years (Landecker, 2016). More recently, resistance has spread and affected the treatment of hospital-acquired infections, such as the Kpn carbapenemase (KPC) in the United States (Venkatachalam, 2012) and the New Delhi metalo-β-lactamase-1 (NDM-1). These genes confer resistance to p-lactam antibiotics and in the majority of cases have been identified in mobile genetic elements that also carry resistance to other antibiotics, such as aminoglycosides and fluoroquinolones (Pendleton, et al., 2013). Recent studies in Colombia have identified an increase in the distribution and dissemination of β-lactamases in Gram-negative bacteria (Rada, et al., 2019), the isolation of Kpn strains harboring the KPC carbapenemase (Hawser, 2009; Jones, 2013; Rojas, 2013), and the high risk associated with carbapenem-resistant strains (Cienfuegos-Gallet, 2019). The spread of resistance associated with transposons in enterobacteria (Cuzon, 2013) and the outbreaks of Kpn strains carrying conjugative plasmids with NDM-1 that can be easily transferred between bacteria have also been reported (Escobar-Pérez, 2013).
The Gram-negative bacterium Kpn can be found in the environment as well as in healthy individuals but is nonetheless an important cause of community and hospital-acquired infections. This microorganism is part of the ESKAPE group of pathogens, which includes Enterococcus faecium, S. aureus, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp., known for their ability to escape the effects of commonly used antibiotics (Pendleton, et al., 2013). It is estimated that Kpn is the cause of a significant amount of hospital-acquired infections in the Western world that can result in hepatic abscesses, septicemia, and respiratory and urinary infections, among others (Shon, 2013). This opportunistic pathogen affects mainly adults, neonates, and immunocompromised patients. However, the emergence of the hypervirulent hvKP strain, capable of infecting healthy individuals and readily acquiring and spreading resistance, has highlighted the risk posed by this pathogen (Lee, 2017). In Colombia, Kpn is one of the microorganisms most frequently isolated from hospitalized adults in intensive care units, as well as in pediatric and neonatal units, with reported increases in the resistance to antibiotics such as carbapenems (Leal, 2016). It was also the most isolated pathogen in healthcare-associated infections between 2012 and 2016 (Instituto Nacional de Salud, 2016).
The global threat posed by antibiotic resistance requires a greater understanding of the conditions that can promote the spread of its determinants via HGT. Recent studies have shown that antibiotics at low concentrations can promote resistance and, in some cases, foster HGT, particularly by conjugation. Concentrations below the minimal inhibitory concentration (MIC) of disinfection agents, such as triclosan, chlorhexidine, chlorine, and hydrogen peroxide, have been shown to affect plasmid conjugation (Jutkina, 2018; Zhang, 2017). Sub-MIC or sub-lethal antibiotic concentrations of β-lactams and tetracycline were shown to increase the transfer of resistance in S. aureus (Barr, 1986). More recently, sublethal concentrations of the antibiotics cefotaxime, ciprofloxacin, and ampicillin were reported to increase the transfer of plasmids bearing resistance genes in Escherichia coli (Liu, 2019). Several antibiotics tested at sub-MIC concentrations also induced plasmid conjugation using diverse bacteria as donors and E. coli as the recipient (Jutkina, 2018; Jutkina, 2016). These studies indicate that sub-lethal concentrations of antimicrobials can increase the conjugation transfer frequency of plasmids bearing resistant genes. Here we study the impact of antimicrobials on HGT using the opportunistic pathogen Kpn as a model to address the hypothesis that exposure to sub-lethal antibiotic concentrations can affect both conjugation and transformation. Diverse types of antibiotics were therefore evaluated for their capacity to affect the acquisition of resistance in bacteria by the transfer of plasmids and by uptake of extracellular DNA, respectively. The results indicate that sub-lethal concentrations of some aminoglycoside antibiotics can increase HGT in this opportunistic pathogen.
Methods
Bacterial strains, plasmids, and growth conditions
Bacteria and plasmids used are listed in table 1. All strains were stored at -80°C in Luria Bertani (LB, oxoid) broth supplemented with 20% glycerol. When necessary, antibiotics (Sigma-Aldrich, St. Louis, MO) were added: Ampicillin (Amp, 200µg ml-1), imipenem (Imp, 2µg ml-1), spectinomycin (Spc, 50 µg ml-1), gentamicin (Gen, 20 µg ml-1), and chloramphenicol (Cam, 20 µg ml-1). Sub-lethal concentrations used were: Amp (2µg ml-1), Amk (0.5 µg ml-1), Gen (0.20 µg ml-1), Kan (0.5 µg ml-1), Spc (0.5 µg ml-1), ciprofloxacin (Cip, 0.05 µg ml-1), Imp (0.25 µg ml-1), and trimethoprim-sulfamethoxazole (TS, 0.004/0,076 µg ml-1). Their growth was monitored by measuring optical density at 595 nm (OD595) and colony-forming units (CFU) were determined by the microdilution technique (Pfeltz 2001). Unless stipulated, strains were grown at 37° C, 180 rpm, in LB or cation-adjusted Mueller-Hinton (MH, Oxoid) broth. Modified LB (7.5 g yeast extract, 15 g tryptone, and 7.5g NaCl in 1 liter dH2O) was used for transformation assays and transformants were selected on LB supplemented with 5% agar and 200µg ml-1 Amp (Zhang, 2012).
Determination of the minimum inhibitory concentration (MIC)
The MIC was determined using 96 well plates (Corning Costar 3595), as reported (Andrews, 2002). Briefly, bacterial cultures grown overnight (o/n) for 16 h were diluted 1/30 and grown for 50 min (37°C at 180 rpm). This culture was then diluted 1/100 in fresh medium to obtain a concentration of approximately 106 CFU/ml; 75 µl of each inoculum were used for assays in the 96 well plates containing 75 µl of varying antibiotic concentrations in cation-adjusted MH broth. The ranges and concentrations of antibiotics were established following previously defined criteria (Clinical & Laboratory Standards Institute - CLSI, 2015). Plates were incubated at 37° C for 16 h without agitation; all assays were done in duplicate. The MIC was defined as the lowest antibiotic concentration at which bacterial growth was inhibited.
Transformation on agar
For natural transformation assays, we followed the previously used protocol (Zhang, et al., 2012) with some modifications. All experiments were done at 30°C; 200 µl of an o/n culture of Kpn CH404 (sensitive to Amp) were inoculated into 20 ml modified LB keeping, the medium to flask volume ratio of 1/5 for aeration, and static incubation was carried out for 24 h; 500 µl were centrifuged, 450 µl of the supernatant were removed, and 50 ng of plasmid DNA (Table 1) were added to the cell suspension. This mix was spread immediately onto LB agar with Amp (200 µg ml-1). When using the antibiotics Cam and Gen, the mix was spread for 3 h on LB before selection with the antibiotic. The frequency of transformation was calculated by dividing the number of transformants by the total number of viable cells determined using the microdilution method (Pfeltz, 2001). To analyze the effect of antibiotics on exogenous DNA uptake, Kpn CH404 was grown in the presence of sub-lethal concentrations of the antibiotics Amk, Amp, Imp, Gen, Kan, Spc, Cip, and TS for 6 h at 37° C before precipitating cells by centrifugation and mixing with the DNA.
Conjugation assays
Bacterial conjugation was done on MH agar as reported previously (Suescún, 2006). The cultures of donor and recipient cells were grown o/n and adjusted to an OD595 of 0.8 and 180 µl and 20 µl of the recipient and donor strains, respectively, were mixed in a 0.2 ml tube; 40 µl of this mix were placed on solid MH with no antibiotic, incubated for 4 h at 37° C, after which cells were recovered and resuspended in 200 µl MH. Serial dilutions were done to determine CFUs using the microdilution technique and plating on solid MH supplemented with sodium azide (150 µg ml-1) and the appropriate antibiotic for selection. E. coli J53 transconjugants were confirmed by purifying the plasmid and by antibiotic resistance profiles. When using E. coli J53 as the donor, Kpn CH404 transconjugants were identified based on morphological characteristics by plating on eosin methylene blue (EMB) agar supplemented with Amp (200 µg ml-1) and Spc (50 µg ml-1) (Holt-Harris, 1916). The antibiotic susceptibility of the recipient and the donor strains were always evaluated to avoid the appearance of false positive colonies. The conjugation frequency was calculated by dividing the total number of transconjugants by the total number of donors. All assays were done in triplicate.
Conjugation in biofilms was done by co-incubating donor and recipient cells to form mixed biofilms on lid pegs of a Calgary device (Thermo Scientific Nunc 445497) (Harrison, 2010). Briefly, o/n cultures were adjusted to 0.5 of the MacFarland standard, then mixed at a 1/10 ratio (donor/ recipient), and 1.5 µl were used to inoculate wells of a Calgary device containing 150 µl MH broth alone or with sub-lethal concentrations of the antibiotics Amp, Amk, Cip, Imp, and TS. After 24 h of incubation at 37° C, the biofilms formed on the lids were washed three times by submerging the pegs in sterile PBS and biofilm cells were removed by a combination of sonication (50Hz) for 3 minutes and vortex for 10 minutes. The resulting suspensions were diluted and plated on selective medium to determine CFUs.
Biofilm quantification
After removing planktonic cells, biofilms formed on lid pegs were washed with PBS, dried at room temperature for 40 minutes, and stained using 0.1% crystal violet (CV) for 20 minutes. Excess stain was removed with distilled water, 250 µl of 10% SDS were added per well and absorbance was measured at 595 nm. All assays were done in triplicate.
Molecular methods
The acquisition of the pUC19 plasmid by Kpn CH404 was confirmed by PCR amplification of the multiple cloning site of approximately 123 bp using universal M13 primers. Plasmid DNA was obtained using the QIAprep Spin miniprep kit (QIAGEN). Plasmids from clinical isolates were obtained following reported methods (Kado, 1981). Restriction enzyme digestion was carried out using XbaI and the products were analyzed by electrophoresis on 1% agarose gels. Chromosomal DNA was obtained using the kit DNA 2000 (Corpogen).
RNA seq and data analysis
An o/n Kpn CH404 was diluted 1/100 in duplicate into 6 ml MH medium with no antibiotic (control) or supplemented with 0.5 ug ml-1 Amk. After 6 h of growth, cultures were centrifuged at 4°C for 10 minutes at 3500 rpm and cells were resuspended in 1 ml TRIzol ™ (Invitrogen™) pre-heated at 65°C. Samples were centrifuged for two minutes at 4°C at 13,000 rpm and the supernatant was mixed by inversion with 1 ml 100% ethanol. The total RNA was recovered using the Direct-zol (Zymo research) kit following the manufacturer's specifications. Only RNA with a RIN >8 was used for sequencing at Macrogen (Soul, Korea) in an illumina Hi-seq 2000 with 100 nucleotide paired-end reads. Quality control was done with FastQC (v.0.11.2) (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/), Illumina adapters were trimmed (Trimmomatic, v.0.36) (Bolger, 2014) and rRNA was removed using SortmeRNA (v.2.1) (Kopylova, 2012) and the Silva16S, 23S and 5S rRNA gene databases (https://www.arb-silva.de) (Pruesse, 2007). The data were analyzed using Rockhopper (McClure, 2013), which calculates differences in expression by determining a p-value (Anders, 2010) and then calculating q values according to the Benjamini-Hochberg correction (Benjamini, 1995) with a false discovery rate (FDR) <1%. This analysis was done on the PATRIC platform (www.patricbrc.org) using as reference the K. pneumoniae subsp. pneumoniae HS11286 genome. Genes with significant differences were considered to be those with a log2 fold-change in expression >2 and a q value <0.001.
Statistical analyses
All experiments were done in triplicate (except for MIC determinations) and the mean ± standard deviation (SD) is shown; a t-Student test for independent samples was used to compare the effect of treatments on HGT. The non-parametric Mann Whitney test was used for non-normal data; significant differences were expressed as p< 0.05. The statistical analyses were done using the software IBM-SPSS version 22.
Results and discussion
Kpn can take up exogenous DNA
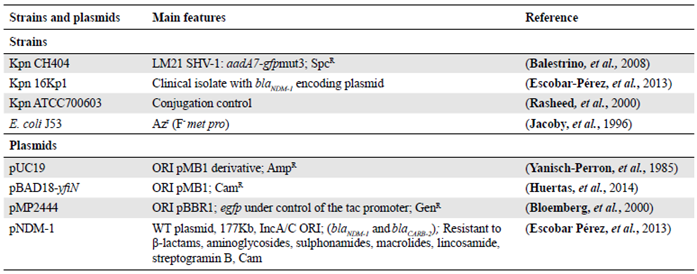
Kpn has genes that are orthologous to those present in naturally-competent bacteria, such as H. influenzae (Blokesch, 2016; Palchevskiy, 2006), yet it is not capable of naturally taking up genetic material from its surroundings. A recent study reported that E. coli, which is phylogenetically close to Kpn and is likewise not considered to be naturally competent, was capable of taking up exogenous DNA under certain laboratory conditions (Zhang, et al., 2012). Therefore, we used this system of transformation on agar to evaluate the capacity of Kpn to acquire exogenous DNA. The laboratory strain Kpn CH404, which is sensitive to Amp, was mixed with the pUC19 plasmid as a source of free DNA and plated on solid medium with antibiotic. As shown in table 2, transformants were obtained, although the frequency was low (approximately 10-10) in comparison to what has been reported previously for E. coli (approximately 10-7) (Zhang, 2012). The transformation efficiency of 104 / µg DNA was also low compared to competent cells prepared in the laboratory to optimize this process (about 106 - 108 transformants/ug of plasmid DNA). To verify the transformation results, plasmid DNA was isolated from AmpR transformed cells and analyzed by restriction enzyme digestion, which revealed a pattern identical to the original pUC19 plasmid (Figure 1). The presence of the plasmid was also verified by amplification of the multiple cloning site in pUC19 using M13 universal primers. No transformants were obtained when the mix (cells + DNA) was treated with DNase I before plating on selective medium nor when the mix was plated on medium containing DNase I, indicating that intact exogenous DNA was required. These results showed that under certain conditions, Kpn is capable of acquiring exogenous DNA.
Table 2 Natural transformation in K. pneumoniae (Kpn)
| Condition | Plasmid | Size (Kb) | Resistance | Origin of | Transformation |
|---|---|---|---|---|---|
| (incubation time) | replication | frequency | |||
| Control (24h) | pUC19 | 2.7 | Amp | pBR322 | (4.7±0.4) x10-10 |
| Control (6h) | pUC19 | 2.7 | Amp | pBR322 | (1.3±0.2) x10-8 |
| Control (6h) | pBAD18-yfiN | 5.8 | Cam | pBR322 | (4.7±0.5) x10-10 |
| Control (6h) | pMP2444 | 5.6 | Gen | pBBR1MCS-5 | (2.9±0.2) x10-10 |
| Free DNA (lysed, 6h) | pUC19 | 2.7 | Amp | pBR322 | (8.3±1.3) x10-8 |
| Sub-lethal Antibiotics | |||||
| *Amk (6h) | pUC19 | 2.7 | Amp | pBR322 | (1.2±0.9) x10-7 |
| *Gen (6h) | pUC19 | 2.7 | Amp | pBR322 | (2.2±0.5) x10-7 |
| *Kan (6h) | pUC19 | 2.7 | Amp | pBR322 | (3.9±0.3) x10-7 |
| Spc (6h) | pUC19 | 2.7 | Amp | pBR322 | (3.3±0.6) x10-8 |
| Amp (6h) | pUC19 | 2.7 | Amp | pBR322 | 0 |
| Cip (6h) | pUC19 | 2.7 | Amp | pBR322 | (2.0±0.8) x10-8 |
| Imp (6h) | pUC19 | 2.7 | Amp | pBR322 | 0 |
| TS (6h) | pUC19 | 2.7 | Amp | pBR322 | (1.7±1.0) x10-10 |
Kpn CH404 cells were grown in modified LB for 24 h or 6 h with or without sub-lethal concentrations of antibiotics: amikacin (Amk) 0.5µg ml-1; gentamicin (Gen) 0.20 µg ml-1; kanamycin (Kan) 0.5 µg ml-1; spectinomycin (Spc) 0.5µg ml-1; ampicillin (Amp) 2µg ml-1; ciprofloxacin (Cip) 0.05µg ml-1; ), imipenem (Imp) 0.25 µg ml-1; trimethoprim-sulfamethoxazole (TS) 0.004/0.076µg ml-1. Transformed cells were confirmed by evaluating antibiotic-resistant profiles, isolation of plasmids, and analysis of electrophoretic mobility on agarose gels and, in some cases, by performing PCR amplification of plasmid sequences. The frequency of transformation was obtained by dividing the number of transformants by the total number of viable cells. The results represent the mean of three biological replicates; asterisks indicate treatments with significant differences vs the untreated control. * p<0.05
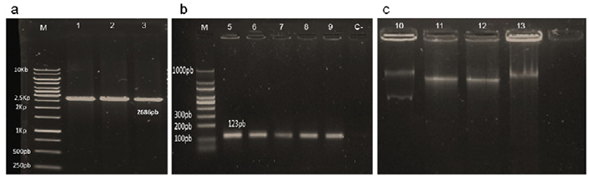
Figure 1 Confirmation of plasmid transfer. Transformation was confirmed by (a) XbaI digestion of plasmid pUC19 (lane 1) and plasmids obtained from transformed cells (lanes 2 and 3), and (b) PCR amplification of the multiple cloning site in pUC19 using universal M13 primers from the plasmid control (lane 5) and transformants (lanes 6-9); C-: negative control. Conjugation was confirmed by (c) pNDM-1 plasmid DNA extraction from Kpn donor strain 16Kp1 (lane 10) and from transconjugants (lanes 11-13). M: DNA Molecular weight markers
To determine if this transformation was limited to this one plasmid, we also evaluated plasmids pBAD18-yifN and pMP2444, that differ in size, origin of replication and the resistance markers Cam and Gen, respectively. In this case, cells were plated on medium lacking antibiotic before using selective medium since these antibiotics inhibit protein synthesis, which has been reported to be required for transport and uptake of exogenous DNA (Sun, 2006). Again, Kpn cells were capable of acquiring both plasmids, although the frequencies were very low (Table 2). We then evaluated if Kpn might take up DNA liberated from lysed cells, which could more closely resemble conditions encountered by this microorganism. When cells bearing pUC19 plasmid were lysed by boiling (95°C for 10 minutes), transformants were obtained at a similar frequency as when using purified plasmid (Table 2). In contrast to these transformation assays on agar, no DNA uptake was observed using other non-conjugative plasmids, pKOBEG199, pMP2444, and pBAD-18, and assays previously reported to result in direct transfer via cell contact (Maeda, 2006)
(Table 2). The use of PCR-amplified linear DNA also failed to give rise to transformed cells, even though in this case recovery of cells bearing the AmpR resistant marker would also have required homologous recombination. The data presented indicate that Kpn can, under very defined laboratory conditions and at very low frequencies, take up exogenous plasmid DNA as has been previously reported for E. coli (Sun, et al., 2006). This uptake may be mediated by competence gene homologs which could allow Kpn cells to acquire novel genes and/or utilize DNA as a source of energy as previously suggested (Blokesch, 2016; Finkel, 2001).
Conjugation increases in biofilms
Given the importance of ARG transfer by conjugation, we next evaluated conjugation using the clinical isolate 16Kpn1 that carries the transferable plasmid pNDM-1. This plasmid contains the bla NDM1 gene that codes for a globally disseminated carbapenemase that confers resistance to carbapenems (Kumarasamy, 2010). Strain 16Kpn1 was isolated from a blood infection at a hospital in Bogotá, Colombia, during the first report of a bla NDM-1 outbreak in the country in 2011 (Escobar-Pérez, 2013). Using conjugation on agar (Suescún, et al., 2006), plasmid pNDM-1 was transferred from 16Kpn1 (pNDM-1) to the recipient strain E. coli J53 (AziR) with a conjugation efficiency of approximately 10-4 (Figure 2). The E. coli transconjugants acquired the resistances present on plasmid pNMD-1 (Amk, Gen, Amp, and Imp) (Escobar-Pérez, et al., 2013) and plasmid preparations indicated the transfer of only one of the two plasmids present in the donor strain (Figure 1). Experiments produced the same results when repeated multiple times. Control reactions lacking donor strains did not give rise to resistant colonies indicating that the observed results were due to transfer of the pNMD-1 by the donor 16Kpn1 and not to contamination. This plasmid was also transferred to strain Kpn CH404 and from this donor strain (Kpn CH404 pNDM-1, AmpR) to E. coli. Finally, the E. coli transconjugant bearing the plasmid (J53 pNDM-1) was also capable of transferring the plasmid to strain Kpn CH404 (sensitive to Amp) indicating that the plasmid has the capacity to be transferred from other bacteria besides Kpn. These results showed that pNMD-1 carrying multiple resistance markers can be transferred via conjugation between Kpn and E. coli and is, therefore, an excellent model for evaluating the effect of antibiotics on conjugation.
Conjugations with different donor-receptor combinations of Kpn (16Kp1 and CH404) and E. coli (E. coli J53) were done for 4 h using planktonic cells or biofilms. The frequency of conjugation was determined by dividing the number of transconjugants by the total number of receptors. The values shown are the mean ± SD of three independent experiments; asterisks indicate treatments with significant differences vs. the untreated control (**p<0.05 and *** p<0.001).
Figure 2 Frequency of conjugation by pNDM-1
Conjugation was next analyzed in biofilms, multicellular structures and ubiquitous forms of bacterial growth in which high rates of plasmid transfer have been observed (Mielich-Süss, 2015). Both the donor and the recipient strains grown separately formed biofilms on the Calgary device, although E. coli J53 formed less biofilm (OD 0.07± 0.02) than Kpn 16Kpn1 (OD 0.51 ±0.06). Co-cultures with both strains resulted in biofilms with an intermediate phenotype (OD 0.24 ± 0.04). Kpn was capable of donating and receiving plasmid pNDM-1 by conjugation in these biofilms with a frequency often greater than in planktonic cells, as shown before (Hoffman, 2005) (Figure 2).
Some antibiotics increase HGT
To analyze the effect of antibiotics on gene transfer, we used five antibiotics with different modes of action: Amp and Imp target the bacterial cell wall, Amk inhibits protein synthesis, Cip inhibits the synthesis of DNA, and TS inhibits folic acid metabolism. The concentrations at which these antibiotics inhibit growth (MICs) were first determined (Table 3) and cell growth was verified to be unaffected in the presence of three different sub-lethal (sub-MIC) antibiotic concentrations tested. Concentrations below the MIC that gave consistent results and did alter growth were selected for assays. The effect of sublethal concentrations of antibiotics on plasmid acquisition by transformation indicated that only the aminoglycoside Amk added 30 minutes before the assay significantly increased the frequency of transformation compared with the control without antibiotic (Table 2). Additional aminoglycosides were therefore tested (Gen, Kan), as well as Spc, which is closely related. Of these treatments only Gen significantly induced transformation compared with the control, whereas Kan and Spc resulted in only a slight increase in uptake of exogenous DNA (Table 2). Together, these results suggest that sub-lethal concentrations of some aminoglycosides can induce uptake of extracellular DNA by Kpn. The fact that only some of the aminoglycosides affected transformation despite having the same mode of action could be due to differences in molecular structure that in turn elicit dissimilar cellular responses and result in diverse responses to the stress imposed by the presence of the antibiotic (Dalhoff, 2018). These changes can, for example, affect the expression or polarity of cell determinants involved in the capacity to interact with the environment or in the acquisition of exogenous DNA.
Table 3 Minimum inhibitory concentrations
| Antibiotic | Kpn 16Kp1 | Kpn CH404 | E. coli J53 |
|---|---|---|---|
| Amp | >400 | 6.25 | 12.5 |
| Amk | 32 | 2 | 8 |
| Cip | 0.016 | 0.0625 | 0.003 |
| Imp | 32 | 0.8 | 0.2 |
| TS | 1/19 | 0.025/0.48 | 0.013/24 |
Data correspond to the lowest concentration (ng/ml) in which growth was inhibited. Both Kpn C0H404 and E. coli J53 strains represent MICs of control plasmid-free strains.
Amp: Ampicillin; Amk: Amikacin; Cip: Ciprofloxacin; Imp: Imipenem; TS: Trimethoprim-sulfamethoxazole
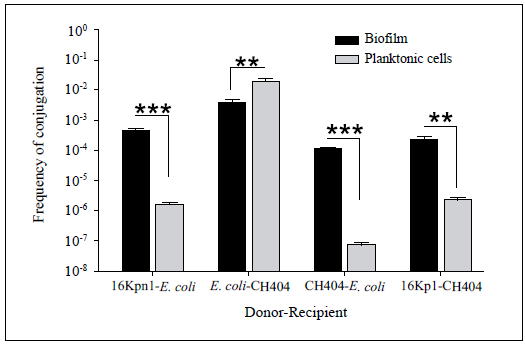
We next evaluated the effect of antibiotics on conjugation using biofilms formed on a Calgary device (see Methods). To measure the effect on conjugation, 16Kpn1 and E. coli J53 cells were grown in the presence of sub-lethal antibiotic concentrations for 24 h to form mixed biofilms. The antibiotic concentrations used did not affect the growth of either the donor or recipient (not shown). Biofilm formation increased almost five times when cells were grown in the presence of Amk (1.32 ± 0.27) and more than two times with Imp (0.81 ± 0.20) compared with the control with no antibiotics (0.28 ± 0.05) (Figure 3a). In the majority of cases, the biofilm was similar for the receptor and donor strains with a notable increase in the amount of adhered donor cells compared with recipient cells in the presence of Amk (black bars, Figure 3b). These results demonstrated that incubation in sub-lethal concentrations of some antibiotics induced biofilm formation in Kpn as has been shown previously for other microbes (Hoffman, et al., 2005). We then determined the number of transconjugants obtained (E. coli recepient of the pNDM-1 plasmid) for each treatment by recovering biofilms and determining CFUs on MH agar medium supplemented with sodium azide and Amp. The frequency of conjugation increased in all treatments but Amk was the only one that showed a significant difference with respect to the control and to the other antibiotics used (Figure 3a). It was interesting to observe that although incubation with Imp increased biofilm formation, there was no significant increase in conjugation, suggesting that forming a more robust biofilm is not necessarily correlated with an increase in DNA transfer via conjugation. This result also indicates that an increased donor:recipient ratio as observed mainly for Amk and Imp (Figure 3b), was not the sole factor affecting conjugation frequency (Figure 3a). It is possible that despite having more cell density and proximity, cells in biofilms do not necessarily have the conditions adequate for expressing the determinants required for efficient conjugation.
These observations were verified by carrying out conjugation on solid media (Figure 3c). In this case, media were supplemented with aminoglycoside antibiotics at sub-lethal concentrations and the donor-recipient mixes were incubated for the same amount of time utilized for biofilm assays on Calgary devices (24 h). Once again, incubation with Amk increased the number of transconjugants approximately ten times. The frequency of conjugation also increased when other aminoglycoside antibiotics were used, with Kan being the one that increased the most (1.4 X 10-1) compared with the control (7.7 X 10-4) (Figure 3c). Cell growth was not affected by these sub-lethal antibiotic concentrations (not shown). Together, these results suggest that some aminoglycosides can increase the transfer of the pNDM-1 plasmid by conjugation, which is the principal mode of transfer for antibiotic resistance markers in Kpn (Skippington, 2011).
(a) Mixed biofilms of donor (16Kpn1) and recipient (E. coli J53) cells formed on lid pegs of a Calgary device in the presence of sub-lethal concentrations of: Amk 0.5 ug ml-1; Amp 2 ug ml-1; Imp 0.25 ug ml-1; TS 0.004/0.076 ug ml-1; Cip 0.05 ug ml-1. The bars show biofilm quantification by CV staining and the dotted line indicates the frequency of conjugation for each treatment (number of transconjugants/total number of receptor cells). (b) Quantification of donor (black bars) and recipient (white bars) cells adhered to lid pegs of a Calgary device. (c) Conjugation on agar in the presence of sub-lethal concentrations of aminoglycosides: Amk 0.5ug ml-1; Gen 0.20 ug ml-1; Kan 0.5 ug ml-1; Spc 0.5ug ml-1. The total number of transconjugants is shown (black bars) per recipient cells (white bars) after 24 h of conjugation (Y-axis, left). Gray bars indicate frequency of conjugation (Y-axis, right). The data represent the mean of three replicas; asterisks indicate treatments with significant differences by CV staining and frequency of conjugation compared to the control (*p<0.05).
Figure 3 Factors affecting conjugation of the pNDM-1 plasmid.
We conducted RNA seq assays to understand the possible mechanism involved in the observed HGT increase caused by sub-lethal concentrations of Amk. Total RNA was isolated from duplicate cultures of controls free of antibiotics and cultures incubated in sub-lethal Amk concentrations (0.5|ig ml-1) for 6 hours. The differential expression analysis carried out using the program Rockhopper (McClure, et al., 2013) on the PATRIC platform identified 367 differentially expressed genes (DEGs) with a significant log2 fold-change (FC) > 2 and a q value < 0.001. Of these genes only one, the KPHS_40620, which codes for an aldehyde dehydrogenase, was overexpressed under the control condition (log2FC = 5.4). The remaining genes (366) had more expression under incubation with Amk (Supplementary table 1, https://www.raccefyn.co/index.php/raccefyn/article/view/985/2689). The majority of the DEGs correspond to functions involved in cellular metabolism (115), followed by transport (65) and regulation (54) (Table 4). Several genes (23) were identified as surface components or proteins involved in secretion. Among the highest DEGs involved in cell metabolism were KPHS_40620 (log2FC = 5.43) and KPHS_41710 (log2FC = 5.25), an aldehyde dehydrogenase and a decarboxylase induced in the control and with Amk, respectively (Supplementary table 1, https://www.raccefyn.co/index.php/raccefyn/article/view/985/2689). The large number of overexpressed genes under Amk incubation that code for cell surface components and proteins involved in transport suggests an effect on cell components involved in sensing or interacting with the environment and/or exchange across the cell membrane. Some of these genes were KPHS_29860 (Type 6 secretion), KPHS_21110 and KPHS_32910 (ABC transporters), rbsB, and oppA, both of which code for surface proteins that have been implicated in conjugation in E. coli (Zhang, et al., 2012). Among the genes identified, KPHS_49140 and KPHS_49350 code for proteins involved in DNA uptake via the type 4 pilus (PilQ) and competence (YhgH), respectively (Blokesch, 2016; Palchevskiy, et al., 2006). We also observed the expression of genes involved in functions that affect cell viability and survival, like KPHS_44010 (secreted protein, sensitivity to copper) and KPHS_03620 (PGA synthesis, biofilms). These results are consistent with previous reports describing changes in the outer membrane of Gram-negative bacteria when they interact with aminoglycosides such as Amk (Dalhoff, 2018). It is believed that these antibiotics alter and permeabilize the bacterial membrane and affect cell viability through electrostatic interactions. In this particular case, the changes at the membrane level could increase the capacity to acquire exogenous DNA. Finally, the induction of various genes involved in regulation indicates that the interaction with Amk also triggers complex signaling cascades within the cell and the regulation of diverse gene products. Some of the regulators that were abundantly expressed included KPHS_29970 (sugar metabolism), KPHS_5285 (YidZ, LysR family), KPHS_50100 (response to metals, NikR) and KPHS_21540 (MarR family involved in resistance) (Supplementary table 1, https://www.raccefyn.co/index.php/raccefyn/article/view/985/2689). However, no induction was seen of capsule genes or of the RpoS sigma factor gene, contrary to previous studies (Baharoglu, 2013; Lin, 2005). This transcriptional regulator has been implicated in competence in E. coli (Zhang, et al., 2012). The observed differences could be due to variations in experimental procedures such as culture incubation conditions, antibiotic concentrations, and exposure time. Finally, it is worth noting that many of the identified genes correspond to hypothetical proteins or proteins of unknown function, which can be expected as the function of a great number of genes identified in sequenced bacterial genomes has not been established.
Table 4 Summary of differentially expressed genes
| Function | Number | Percentage (%) |
|---|---|---|
| Hypothetical/Unknown | 86 | 23 |
| Metabolism | 115 | 31 |
| Transport | 65 | 18 |
| Regulation | 54 | 15 |
| Surface/Secreted | 23 | 6 |
| Recombination/Repair | 8 | 2 |
| Mobile elements | 4 | 1 |
| Toxin/Antitoxin | 3 | 1 |
| Stress | 2 | 1 |
| Others | 7 | 2 |
| Total | 367 |
The exposure of cells to sub-lethal antibiotics has been reported previously to alter gene expression in diverse microorganisms (Andersson, 2014). Consistent with our results, a recent report indicated that sub-lethal concentrations of carbapenems also affected the expression of genes involved in metabolism, signal transduction, and cell wall processes in Kpn grown as biofilms (Van Laar, 2015). Protein synthesis inhibitors at sub-lethal doses have been shown to result in varied transcription profiles, depending on the antibiotic tested (Lin, et al., 2005). In our case, sub-lethal amikacin (Amk) resulted mostly in the induction of genes. These genes correspond to diverse functions and are not necessarily in operons or known regulons suggesting that the effect is not just due to the inactivation of regulators of gene expression. The high number of DEGs identified as being involved in cell envelope and extracellular functions, as well as genes involved in recombination and repair, correlate with previous studies on genes expressed due to sub-lethal antibiotic concentrations (Shun-Mei, 2018; Van Laar, et al., 2015) and suggests that some of these might be good candidates for additional analysis. RNA seq has the advantage of providing a transcriptional profile that does not rely on previous sequence information, yet the complexity of the analyses and the large number of genes involved in any given cellular response also limit the interpretations. Thus, understanding the precise role of the genes identified and their probable involvement in HGT requires further experimental validation.
Our study showed that exposure to sub-lethal antibiotic concentrations affects HGT by transformation and conjugation of mobile genetic elements, some of which can carry multiple resistance genes. Thus far, very little is known regarding signals like temperature, oxygen, nutrients, pH, and osmolality capable of inducing conjugation (Frost, 2010), especially in clinical isolates. Here, sub-lethal aminoglycoside concentrations could enhance HGT, apparently by inducing cellular changes involved in shaping the cell envelope, metabolism, interaction with the environment, and regulation. Although additional work on particular gene products would be required to confirm these observations, the identification of factors that affect HGT in bacteria such as Kpn provides critical information regarding the effect of sub-lethal antibiotics concentrations on gene transfer that may affect the spread of resistance in hospital settings.
Conclusions
Sub-lethal concentrations of antibiotics present in diverse environments can favor the appearance of resistance to these drugs and the spread of ARGs by HGT (Andersson, et al., 2014). This study indicates that some aminoglycosides, in particular Amk, can affect transformation and conjugation in the opportunistic pathogen Kpn, a significant cause of hospital-acquired infections. RNA seq assays suggest that sub-lethal concentrations of Amk alter gene expression and components of the cell envelope, processes that could modify the cell's capacity to acquire genetic material via HGT. Additional confirmatory experiments would be required to fully understand the molecular mechanisms involved and the extent of the effect of these antibiotics on HGT. Understanding the conditions that influence the acquisition and spread of mobile genetic elements and resistance to antibiotics can raise awareness regarding the potential effects of antibiotics on the spread and persistence of resistance determinants and bacterial pathogens such as Kpn that pose a risk to human health.