Introduction
Paracoccidioides spp. is a genus of temperature-dependent dimorphic fungi that causes paracoccidioidomycosis (PCM), a neglected tropical systemic mycosis. Brazil reports 80 % of the cases with an estimated incidence of 3 to 4 new cases/million and up to 8 to 10 cases/million in highly endemic areas (Bellissimo-Rodrigues et al., 2011; Coutinho et al., 2015). Since 1908, over 15,000 cases have been reported. The mycosis exhibits two main clinical presentations: the subclinical, or asymptomatic infection, and the clinically manifested disease. The latter is usually chronic and involves the primary target, i.e., the lungs, as well as the mucosa and different organs such as the skin, the adrenal glands, and lymph nodes, among others. PCM affects adult males at least ten times more than women, with a male-female ratio of approximately 11:1 (Restrepo-Moreno et al., 2020). The disease has long latency periods, up to 30 years in some cases, as evidenced by cases reported outside the endemic area (Restrepo, 2000).
The first two cases of PCM were reported in 1908 by Adolpho Lutz (1855-1940) (Lutz, 1908, 2004). In 1912, Alfonso Splendore (1871-1953), a colleague of Dr. Lutz, reported four new cases of the disease and placed the fungus in the genus Zymonema, as Zymonema brasiliensis (Splendore, 1912). In 1930, Floriano Paulo de Almeida (1898-1977) published a comparative study of Coccidioides immitis and this new agent demonstrating that PCM and coccidioidomycosis were two different diseases and classifying the agent as Paracoccidioides brasiliensis (Almeida, 1930). A consensus on the name and classification was finally achieved in 1971 at the PCM symposium in Medellin, Colombia (PAHO, 1971).
Initially, the disease was named Brazilian blastomycosis, then South American blastomycosis, Lutz disease, and Lutz-Splendore-Almeida disease, among other names. Today, PCM is the universally accepted denomination (Franco et al., 1994).
Since its discovery, Paracoccidioides has been the focus of intense research, but despite the exquisite characterization of the immune response and the biochemical characteristics of Paracoccidioides, the ecology and evolutionary biology of the fungus remained largely unexplored until the advent of molecular biology. The use of gene genealogies and later the use of wide phylogenetic reconstructions of the genome revealed the existence of species boundaries within what was previously thought to be a single species. The genus Paracoccidioides now encompasses P. lutzii, P. brasiliensis sensu stricto, P americana, P. venezuelensis, and P restrepiensis. The five species show phenotypic differences in their morphology (Turissini et al., 2017), pathogenesis (Scorzoni et al., 2018), and the immune response elicited in the host (Assolini et al., 2021; Siqueira et al., 2016), although there has been no systematic assessment of the phenotypic differences among all species.
Notably, the species have distinct yet overlapping geographic ranges suggesting that opportunities for hybridization and gene exchange do exist (Matute et al., 2006) The most prevalent species in Colombia was named P restrepiensis in honor of Angela Restrepo, a trailblazer in the study of the biology of the fungus (Turissini et al., 2017). All the PCM cases reported in Colombia and subjected to molecular characterization have been classified as P. restrepiensis suggesting that this species is particularly important in Northern South America (Matute et al., 2006).
The extent of genetic variation in Paracoccidioides has been studied since the advent of molecular biology techniques in the 1990s. Initial surveys with RAPDs and RFLPs revealed variations but didn't ascribe them to species boundaries (Morais et al., 2000; Nino-Vega et al., 2000; Soares et al., 1995). The first formal genetic efforts to characterize the extent of genetic variation in P. brasiliensis sensu lato populations used Sanger sequencing to detect genetic polymorphism in short amplicons and revealed the existence of four genetically isolated clades that satisfied the definition of phylogenetic species. Later, these approaches revealed the existence of five different genetically isolated clades that satisfied the definition of phylogenetic species (Matute et al., 2006; Teixeira et al., 2009; Turissini et al., 2017).
Genome sequencing and population genomics allowed for more formal speciation tests (Matute & Sepulveda, 2019). However, the study of short sequences is inherently limited because it can only reveal the history of individual genes while the incorporation of genome-wide polymorphism has provided the opportunity of collating multiple gene genealogies and extracting the most likely species tree. This approach could be used to detect species boundaries using genome divergence as a metric of the concordance between different loci in the genome. In cases of advanced speciation, a large proportion of the genome should show consistent phylogenetic patterns suggesting in turn sufficient time to accumulate private genetic differences (Schön et al., 2009), purge ancestral polymorphism (Gao et al., 2015; Guerrero & Hahn, 2018), and reduce the chance of gene flow (Dagilis et al., 2022; Hamlin et al., 2020). Mavengere et al. (2020) used 37 genomes in genome-level tests for genetic divergence (Mavengere et al., 2020) confirming that the five phylogenetic species within Paracoccidioides fulfill all these requirements.
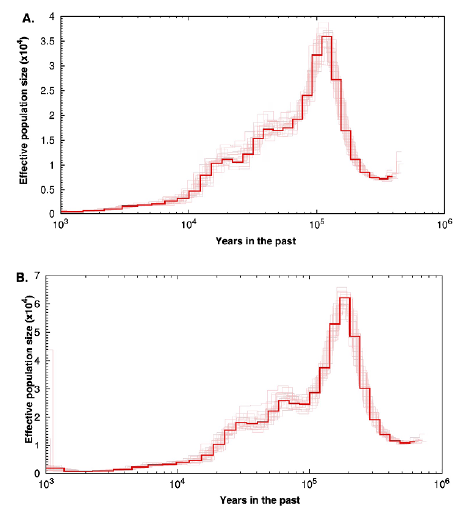
Previous assessments of genetic diversity have suggested that Paracoccidioides species have differences in their polymorphism level (Matute et al., 2007; Mavengere et al., 2020; Turissini et al., 2017) but no study has addressed whether the demographic trajectories of Paracoccidioides species differ from each other. To this end, we used the pairwise sequentially Markovian coalescent (PSMC) method to compare the demographic patterns of two Paracoccidioides species: P. restrepiensis and P. brasiliensis sensu stricto. Our results suggest that, as previously proposed, P. restrepiensis has a lower population size as a consequence of a rapid decrease in its population size and genetic variability compared to P. brasiliensis sensu stricto. Here we speculate on the potential reasons behind this pattern of genetic variability.
Methods
Genomic data
To infer the changes in effective population size, we used the pairwise sequentially Markovian coalescent (PSMC) (Li & Durbin, 2011) method, which uses the genome of a single diploid individual per species. Since all species of Paracoccidioides are haploid, PSMC cannot be applied directly to the genome of a single isolate. Instead, we used two haploid individuals to generate a pseudodiploid genome. We generated two pseudodiploids for P. restrepiensis, one for P. brasiliensis sensu stricto, and one for P. americana. Although our focus was P. restrepiensis, the pseudodiploids from the two other species allowed us to compare it to the two other species. The Sequence Read Archive (SRA) numbers for the genomes used to generate pseudodiploids are listed in table 1.
Read mapping and variant calling
We mapped reads from the six resequenced genomes of the five known species to the P brasiliensis strain Pb18 genome (BioProject accession number PRJNA28733 and BioSample accession number SAMN02953720) using the Burrows-Wheeler Aligner (BWA) version 0.7.12. This assembly currently has 57 supercontigs (Desjardins et al., 2011). The resulting BAM files were merged using SAMtools version 0.1.19 for the variant call step. We remapped reads locally in the merged BAM files using the GATK version 3.2-2, RealignerTargetCreator, and IndelRealigner functions (DePristo et al., 2011; McKenna et al., 2010) and identified indels. Next, we identified single-nucleotide polymorphisms (SNPs) using the GATK UnifiedGenotyper function with the parameter "het" set to 0.01 and all others left as default. (The low het setting obeys to the fact that Paracoccidioides is haploid.) As in previous studies (Mavengere et al., 2020), we filtered the Variant Call Format (VCF) file with the following parameters: QD = 2.0, FSfilter=60.0, MQfilter = 30.0, MQ_Rank_Sum_filter = -12.5, and Read_Pos_Rank_ Sumfilter = -8.0. We excluded sites when their sequencing individual coverage was below the 5th quantile or above the 99th quantile of the genomic coverage distribution from any analyses.
Phylogenetic tree
To infer divergence times between P. restrepiensis and its sister species P. venezuelensis, we used a genome-wide phylogenetic tree which inferred the genealogical relationships between the five known species of Paracoccidioides using a concatenated matrix of polymorphic sites. To convert tree distances to years, we used the mutation rate measured for Saccharomyces in mutation accumulation lines (Leffler et al., 2012), an approach consistent with Turissini et al. (2017).
Generation of pseudodiploids
PSMC uses levels of heterozygosity to infer population changes over time. Since Paracoccidioides is haploid, the method cannot be applied directly to assembled genomes. We used an alternative approach combining data from two individuals to generate a pseudo-diploid. To create pseudo-diploid sequences, we randomly selected heterozygous alleles from haploid genomes of two isolates from a sample of previously sequenced Paracoccidioides genomes (Mavengere et al., 2020). We created pseudo-diploid sequences using the mergefa program in the seqtk package (https://github.com/lh3/seqtk) by merging haploid sequences of two individuals per species (Table 1). We only included sites with coverage between the 5th and 99th quantile of the genomic coverage distribution for both haploid isolates and if the SNP failed to pass one of the GATK filters.
PSMC
PSMC calculates the effective population along slices of time using information from the rates of the coalescent events at a given time. To do this inference, we used the method developed by Li & Durbin (2011) (https://github.com/lh3/psmc). Briefly, the method uses the distribution of heterozygote sites across the genome and the pairwise sequentially Markovian coalescent (PSMC) method that defines a hidden Markov model where the parameters are the mutation rate, the recombination rate, and the effective population sizes through time. The parameters are inferred through an expectation-maximization algorithm. To assess the robustness of the inference, we did 100 bootstraps for each Paracoccidioides species. It should be noted that, unlike standard diploid PSMC plots, PSMC plots of pseudo-diploid genomes usually show an infinite effective population size at the point of divergence (Bazzicalupo et al., 2022; Sato et al., 2020). To convert generations to years, we used a substitution rate for nuclear DNA of 1 x 10-9 substitution/site/year following (Farlow et al., 2015; Lynch et al., 2008).
Results and comments
Paracoccidioides restrepiensis diverged from its sister species recently
Turissini et al. (2017) used a multilocus sequence typing approach to infer the approximate divergence time of Paracoccidioides species. We expanded this analysis using the genome-wide phylogenetic tree inferred by Mavengere et al. (2020)(Figure 1). We found that the approximate age of divergence between P restrepiensis and P. venezuelensis is 125,000 (± 42,000) years, which is consistent with that inferred from a handful of nuclear loci and a constant substitution rate (Turissini et al., 2017).
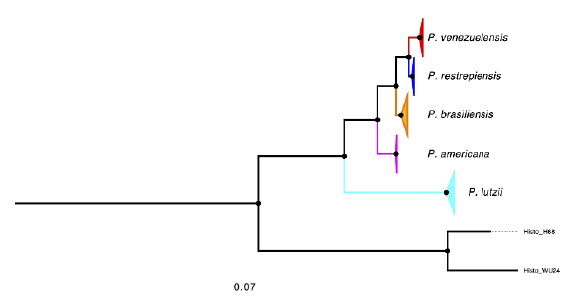
Figure 1 Maximum likelihood tree depicting the genealogical relationships between Paracoccidioides species. Paracoccidioides venezuelensis is shown in red, P. restrepiensis in blue, P. brasiliensis in orange, P. americana in pink, and P. lutzii in cyan. All branches are supported by a bootstrap > 70%. Redrawn from Mavengere et al (2020).
Paracoccidioides restrepiensis has undergone a systematic population bottleneck
Previous analyses have shown that P. restrepiensis has a low level of genetic variation suggesting the possibility of a population bottleneck at some point in its evolutionary history. This pattern of low variability is consistent across the whole genome (Mavengere et al., 2020). We investigated the trajectory of the effective population size in this species using PSMC in two different pseudodiplod genomes. The analyses showed a systematic reduction in the effective population size (TV) over the last 100,000 years (Figure 2); currently, the species shows its lowest historic Ne. This reduction in the N e has a similar timeline to the divergence time from P. venezuelensis (Turissini et al., 2017), P. restrepiensis sister species. Please note that although these dates are heavily affected by our assumptions of mutation rate and the number of generations per year, the pattern of constant Ne reduction remains unchanged.
Paracoccidioides brasiliensis sensu stricto and P. americana
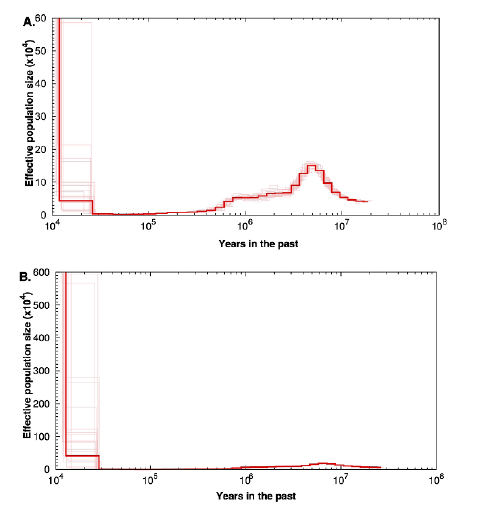
We used similar PSMC analyses with two other species of Paracoccidioides to assess if the pattern of N e reduction over time was unique to P. restrepiensis or whether other species showed a similar trajectory. The analyses yielded two results that are noteworthy: First, we observed that for these other two species, the current effective population size is much larger than that of P. restrepiensis, which is in line with previous observations based on the level of heterozygosity (Mavengere et al., 2020; Muñoz et al., 2016; Turissini et al., 2017). The higher level of these two species' genetic diversity is a historical trend and the two species have had a larger N e than P. restrepiensis for at least the last million years. Second, neither of these two species shows the dramatic effective population size reduction observed in P. restrepiensis. The different trajectory of P. restrepiensis compared to that of the other Paracoccidioides species poses the question of the nature of biogeo-graphic events that led to such a dramatic difference (Figure 3).
Discussion
The evolutionary history of the different Paracoccidioides species remains a largely unstudied realm of the biology of the fungus. To date, most of the research has focused on describing the phylogenetic relationships between the different Paracoccidioides clades (Matute et al., 2006; Teixeira et al., 2009; Teixeira et al., 2014). In this report, we focused on the evolutionary dynamics within Paracoccidioides species and the trajectory of the Ne in P. restrepiensis. Our results suggest that P. restrepiensis, unlike other Paracoccidioides species, has undergone a continuous reduction of effective population size, which ultimately seems to have led to a genetically depauperate extant population.
Our results open two new research avenues regarding the biology of P. restrepiensis. First, recent collections suggested that P. restrepiensis is found not only in Colombia (Cocio et al., 2020; Mattos et al., 2021; Teixeira et al., 2020), which represents a formal test of the existence of isolation in the potential face of gene flow. More importantly, the expanded geographic range needs to be explored from the population genetics point of view. Multiple approaches have used the algorithm STRUCTURE to detect genetic partitions in the Paracoccidioides genus (Bagagli et al., 2021). These analyses have revealed the existence of genetic clusters consistent with the species delimitations from phylogenetic approaches. Nonetheless, they have not detected any population differentiation with P. restrepiensis. This absence of differentiation is not caused by the lack of power to detect structures within each of the phylogenetic species. For example, STRUCTURE detected the existence of two different clades of P. brasiliensis sensu stricto, S1a and S1b (Munoz et al., 2016). Genome-wide tests for speciation revealed that the magnitude of genetic differentiation, albeit considerable, is not sufficient to classify these clades as independent species (Bagagli et al., 2021; Mavengere et al., 2020). The recent collection of P. restrepiensis in southern Brazil poses the question of whether this lack of structure and low historic effective population size are artifacts of focal collection in Colombia or whether the species is truly well mixed (Teixeira et al., 2020).
One of the questions that arise concerns the sources of variation in P. restrepiensis because populations that have undergone extreme bottlenecks are more prone to extinction than those with large effective population sizes. Using full genome sequences and an array of statistical genetics, phylogenetics, and population genetics methods, previous studies have determined that P. restrepiensis evidences the exchange of a moderate number of alleles with other Paracoccidioides species similar to the levels observed among other Paracoccidioides species pairs. These observations are puzzling because the total amount of introgressed DNA was similar across the three species pairs and, given the different ages of the dyads and the expectations, there should be more hybrid incompatibilities in more diverged species pairs than in those recently diverged and, consequently, less possibility of introgression (Dagilis et al., 2022). It is also puzzling that the amount of total introgression in these species pairs was comparable to that observed between species of the genus Coccidioides (C. posadasii and C. immitis), which diverged over 5 million years ago (Maxwell et al., 2019). Whether P. restrepiensis can survive through a constant influx of introgression or through evolutionary rescue, "defined as the recovery of a population through natural selection", is a question that needs to be addressed with a systematic approach.
Genetic divergence is an inexorable outcome of the passage of time. The recognition of species (using any of the available concepts) is not simply a scholastic exercise. On the contrary, the identification of species boundaries allows for the systematic exploration of clinical and epidemiological features that might distinguish between lineages. The use of distinct taxonomic names is also not set in stone. Names have fallen into disuse before (Lacaz et al., 1997). The decision of giving a name to P. restrepiensis was not taken lightly; we studied the extent of differentiation at the genome level, at diagnostic loci (Matute et al., 2006) and in morphological traits (Teixeira et al., 2014; Turissini et al., 2017). We also measured the extent of gene flow between species and found no evidence of introgression, a mark of an advanced level of speciation. The only stronger metric of speciation would be the measurement of reproductive isolation in controlled environmental conditions in a systematic crossing scheme. This experiment, however, is not possible for Paracoccidioides. The fungus is not known to reproduce sexually in laboratory conditions (even though the genome shows evidence of recombination in nature) (Matute et al., 2006). Doing crosses between different species tends to be much more difficult than setting up crosses within species, a prospect that in Paracoccidioides is just staggering.
In the case of P. restrepiensis, there is no evidence that the fungus causes a disease that fundamentally differs from that caused by other Paracoccidioides species, although no study has sought such differences. These studies are cumbersome and will require a systematic assessment of a disease that is characterized by its long incubation period (Restrepo, 2000; Restrepo-Moreno et al., 2020). Nonetheless, there are indications that P. restrepiensis differs from other species in clinically important traits. Serum IgG and IgE show different levels of reaction to antibody exposure in P. lutzi, P. americana, and P. restrepiensis (Assolini et al., 2021). Similarly, even before the species boundaries were reported, P. restrepiensis was reported to have an unusually high level of antifungal resistance compared to isolates subsequently found to belong to the other Paracoccidioides species (Restrepo & Arango, 1980). The generality of these findings at the species level will require a full assessment of the phenotypic trait range across species.
Currently, there are few medically approved drugs for the treatment of mycoses; besides, the drugs have also been used for agricultural purposes for decades. The genetic information of pathogens is dynamic and they are constantly adapting to new hosts. Fungi such as Paracoccidioides spp. have highly efficient machinery (mutations, recombination, and horizontal gene transfer, among others) for genetic diversification and colonization of new environments (Taylor et al., 2017). Emerging genomic variants associated with the antifungal resistance of other fungal species are being reported all over the world. Now, P. restrepiensis is a unique pathogenic fungal species circulating almost exclusively in Colombia and, therefore, it is a priority to continue with the genome surveillance and evolutionary history reconstruction of this endemic fungal species, particularly in a country with high biodiversity and active agricultural production as is the case of Colombia.