Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Colombia Médica
On-line version ISSN 1657-9534
Colomb. Med. vol.44 no.1 Cali Jan./Mar. 2013
Topic review
Functional differences of Porphyromonas gingivalis Fimbriae in determining periodontal disease pathogenesis: a literature review
"Prevalencia de los genotipos FimA de Porphyromonas gingivalis en diferentes poblaciones mundiales: Revisión de la Literatura"
Moreno, Sandraa; Contreras, Adolfob
aEscuela de Odontología de la Facultad de Salud de la Universidad del Valle, E-mail: odsandramoreno@hotmail.com.
bGrupo de Investigación Medicina Periodontal de la Universidad del Valle, E-mail: adolfoco@yahoo.com.
Abstract
Porphyromonas gingivalis is implicated in chronic and aggressive periodontitis. This bacterium has numerous virulence factors and one is the Fimbriae, which is quite important for bacterial colonization. Fimbriae are appendices that anchor to the bacterial wall and are comprised of the protein FimBriline encoded by the FimA gene. Thus far, six genotypes have been identified, FimA I to V and Ib. Genotypes II and IV are associated with periodontal disease, while genotype I is related to gingival health. Genotype identification of P. gingivalis FimA in periodontitis would be important to confirm the pathogenic genotypes and to establish risk at population level. This review is about the P. gingivalis FimA genotype prevalence worldwide. A systematic search using Pubmed, Hinary, and Science Direct within the following descriptors: Porphyromonas gingivalis, bacterial adhesion, periodontitis, Fimbriae, FimA, genotipification was performed to April 2011.
Keywords: Porphyromonas gingivalis, bacterial adhesion, periodontitis, Fimbriae, FimA, genotipification.
Resumen
Porphyromonas gingivalis es un microorganismo implicado en la periodontitis crónica y agresiva. Dentro de sus factores de virulencia, se encuentran las Fimbrias, las cuales están compuestas por una proteína denominada FimBrillina, que está codificada por el gen FimA, del cual existen 6 genotipos (I, II, III, IV, V, Ib), según la secuencia de nucleótidos. Los genotipos II y IV han sido relacionados con periodontitis, mientras el I con salud periodontal. Identificar los genotipos de FimA de P. gingivalis en pacientes con periodontitis podría generar nuevas estrategias que conlleven a suprimir los genotipos más patogénicos para prevenir el desarrollo de la periodontitis en portadores sanos. Se revisó la prevalencia de los genotipos de FimA de P. gingivalis en diferentes países del mundo, para lo cual se realizó una búsqueda sistemática en bases de datos de Pubmed, Hinary y Science Direct usando los descriptores: Porphyromonas gingivalis, adhesión bacteriana, periodontitis, Fimbrias, Fim A, y genotipificación hasta abril del 2011.
Palabras Clave: Porphyromonas gingivalis, adhesión bacteriana, periodontitis, Fimbrias, Fim A, genotipificación.
Introduction
Periodontitis is an inflammatory disease that affects the protective connective tissue, the sulcular epithelium, and the supporting tissue of the teeth like the periodontal ligament and the alveolar bone 1,2.
One of the disease's triggering factors is the persistence of the biofilm formed over the dental surfaces and in the subgingival environment3, which stimulates the immune response of the host and induces the production of proinflammatory cytokines and lysosomal enzyme release by macrophages, besides reduced collagen synthesis by fibroblasts and increased metalloproteinases of connective tissue with concomitant reduction of their inhibitors. This unbalance starts the destruction of periodontal tissue, including bone resorption due to the chronicity of the microbial and inflammatory challenge.4
Diverse microorganisms have been implied in the pathogenesis of periodontitis, like: Porphyromonas gingivalis, Prevotella intermedia, Tannerella forsythia, Fusobacterium nucleatum, Aggregatibacter actinomycetemcomitans, Micromonas micra, Eikenella corrodens, Treponema denticola,and Campylobacter rectus, among other species5.
These microorganisms must find an appropriate ecological niche or activity site in the host to establish themselves and start to grow and multiply, in such a way that they penetrate the physical barriers like the junctional and surcular epithelium, degrade the immunoglobulins in the crevicular fluid and in the saliva, and survive the local action of polymorphonuclear leukocytes (PMN), to then associate with the epithelial cells, cells of the basal layer of the epithelium, and probably the fibroblasts. This association or bacterial adherence, essential for colonization and pathogenicity, is established through the virulence factors that like the Fimbriae facilitate colonization, maintenance, and protection of the bacterial species in the host.6
For Porphyromonas gingivalis (P. gingivalis), the virulence factors are: the capsule7, proteins of the outer membrane, lipopolysaccharides6, proteases like gingipains, collagenases, hemolysin, trypsin proteases, hemagglutinins8, and Fimbriae, which are involved in colonization, invasion, establishment, and persistence within the host, evasion by the destruction of the mechanisms of the immune system and damage to protective periodontal tissues.6,7
This literature review (Pubmed, Hinari and Science Direct to April 2011) emphasizes on one of the virulence factors of Porphyromonas gingivalis such as Fimbriae encoded by the FimA gene, its prevalence in different world populations and the clinical importance from its distribution in the stages of periodontal disease. A systematic search was conducted using the health descriptors: Porphyromonas gingivalis, FimA, genotipification, colonization, and periodontitis in Pubmed, Hinary, and Science Direct databases.
Porphyromonas gingivalis
P. gingivalis is an anaerobic, non-motile, Gram negative coccobacillus, which is related to the start and progression of the chronic and aggressive periodontal disease; it can be considered one of the main etiologic agents of destructive periodontal disease9,10, although it is a microorganism that has also been isolated in gingivitis and in healthy patients in low proportions.7
Likewise, it has been implied in diverse systemic complications like cardiovascular disease, preeclampsia, and low birth weight10,11, given its capacity to colonize other tissues, which has been evidenced by its presence in atheromatous plaques.1.
According to the model of the dental biofilm complexes proposed by Socransky and Hafajee, P. gingivalis belongs to the red complex; hence, it is part of the tertiary colonizers that colonize dental and periodontal tissue when the biofilm is mature.3
This colonization is influenced by saliva, which functions as a vector for its transmission and entry into the oral environment; additionally, the film acquired of the saliva on the dental surface facilitates adhesion of the bacteria Fimbriae, which bond to solid surfaces with high affinity hindering its removal by the salivary flow.1 Finally, these are saccharolytic bacteria with proteolytic activity.
In Vitro, when growing in agar supplemented with sheep blood, hemin and menadione in incubation in anaerobiosis for 10 to 15 days, the colonies are initially observed as yellow or brown colored and after 4 to 8 days they darken from the border to the center until turning black due to their capacity to attract iron from the culture medium12 (Figure 1).
Fimbriae from Porphyromonas gingivalis
Among the virulence factors that develop p. gingivalis, there are Fimbriae, which have been considered the main virulence factor of this microorganism, given that it confers it the capacity to adhere and invade tissues, which characterizes its high pathogenicity over periodontal tissue.9
Fimbriae are fine and numerous appendices that protrude from the outer cell membrane, whose main function is adhesion to periodontal tissue12, endothelial cells1, and other tissues8, given that they have isolated from ovarian and lung abscesses10, and it has even been found that P. gingivalis Fimbriae mediate the congregation with Streptococcus oralis through molecules, residues, or specific domains.13
Two types of Fimbriae exist, the 41-KDa (major) determined by Yoshimura et al., in 1984, and the 67-KDa (minor) found by Hamada et al., in 1996.14. The major, which configure long appendices measuring approximately 0.3 to 1.6 micra long15, is comprised of subunits of a protein called FimBriline, which is encoded by a gene denominated FimA14 of which only one copy exists in the P. gingivalis16 chromosome. Minor Fimbriae are comprised of minor Fimbria protein subunits (Mfa1) encoded by the mfa1 gene14; these Fimbriae measure from 3.5 to 6.5 nanometers long, significantly shorter than the major Fimbriae.15
In research on mice, it has been found that Fimbriae stimulate the production of interleukin 1 (IL-1), interleukin 6 (IL-6), interleukin 8 (IL-8), and tumor necrosis factor (TNF) by peritoneal macrophages and that in humans it triggers TNF secretion per monocyte6. These cytokines are potent inflammatory mediators, which can lead to the activation and destruction of bone tissue and periodontal tissue. Hence, Fimbriae, besides being considered important virulence factors in colonization and invasion of P. gingivalis in oral tissues, can also collaborate with the inflammatory response dependent on the immune response upon stimulating secretion of the cytokines mentioned.
Studies in experimental animals have shown that immunization with P. gingivalis FimBriline protects the individual from destruction of periodontal tissue, but due to the genotypic diversity observed in this bacteria structural and antigenic heterogeneity exists of the Fimbriae, in such a manner that if antibodies are developed against a specific type of Fimbriae protection is not accomplished against the other types.17.
Genotypic and phenotypic variety of the Fimbriae
P. gingivalis is a microorganism with considerable genotypic diversity; hence, we can find clones more pathogenic than others and this could be the reason that explains the presence of the bacteria in healthy patients who have no signs of periodontal disease and in patients with severe periodontal disease, where there are signs of marked destruction of supporting tissue.
FimA is the gene that encodes the Fimbriline subunits. Until now, six FimA genotypes have been found (I, Ib, II, III, IV, V) based on their nucleotide sequence.8, 12 FimA Fimbriae adhere to different proteins of eukaryotic cells like fibronectin, collagen, laminin, the proline-rich protein derived from saliva and statherin, as well as to prokaryotic proteins like glyceraldehyde-3-phosphate dehydrogenase (GAPDH) of Streptococcus oralis.
The FimA gene is found in a gene cluster that encode for regulatory factors or Fimbriline accessory proteins. Downstream from FimA there are four genes denominated FimB, FimC, FimD, and FimE. Recently, it was described that FimC, FimD, and FimE products playa n important role in Fimbriae, in that if they are suppressed in in vitro experiments, the ability of P. gingivalis to adhere to eukaryotic and prokaryotic cells, which affects the capacity of P. gingivalis Fimbriae to colonize tissues, affecting autoaggregation.15
Nagano et al., in 201018, conducted a study with the FimB gene and concluded that this gene regulates FimA length and expression, which is important for colonization and adhesion of the microorganism and generated the hypothesis that the FimB gene would be implied in producing an element responsible for the anchoring of Fimbriae to the bacterial outer membrane.
Prevalence of FimA in diferent world populations
Genotypic variability among P. gingivalis isolates is a reflection of the distinct sequences of its DNA, for this reason the molecular tipification methods have been a big contribution for its study.19. In specialized literature, it is possible to find various studies that have determined FimA prevalence in different world populations (Figure 2).
Because of this, a literature review must be conducted; a dendrogram was carried out in this manuscript to determine population differences through the geographic distribution of said gene, in P. gingivalis (Table 1).
In Japan, Amano et al., have done extensive research of P. gingivalis Fimbriae and their distribution and prevalence in that country's population. In a study in 1999,7 they found that FimA II followed by FimA IV are the most prevalent genotypes in patients with periodontal disease. In that study, they selected 93 Japanese adults with periodontal disease and without systemic involvement (33 men between 19 and 74 years of age and 60 women between 21 and 79 years of age). It should be mentioned that in 1999 only four 4 FimA genotypes were known (from I to IV). The results demonstrated that of 93 patients, 73 were positive for P. gingivalis, of which FimA type II was the most prevalent with 58.9%, compared to 5.4%, 6.8%, and 12.3% for type I, III, and IV, respectively. They also observed a relationship between FimA II with the depth of the pocket, given that in the deepest 90.9% was found, compared to the other genotypes studied, and that in women aged between 36 and 57 years the highest percentage of 83.3% was found for FimA II. Thereby, Amano et al., 1999, concluded that the FimA II genotype is most prevalent in patients with periodontal pathology and that the probability exists of other genotypes different from genotypes from I to IV, which is why they recommend conducting further studies.
In 2000, Amano et al.20 managed to clone a new FimA genotype, the type V. Given that in prior studies some patients who were positive for P. gingivalis had none of the four genotypes known till then; it was suspected that another non−described genotype existed. Of the 73 samples used in the previous study, five specimens had the non-described genotype; hence, these samples were used as templates to amplify the new FimA gene by using a PCR test. The PCR products were separated via electrophoresis and the DNA was cloned in a vector (E. coli).
Later, in 2000, Amano et al.21 studied 380 periodontally and systemically healthy Japanese adults (163 men between 30 and 70 years of age − mean of 51.23 ± 11.2 years of age − and 217 women between 30 and 70 years of age) and 139 Japanese adults with periodontitis and without systemic involvement (70 men between 30 and 70 years of age and 69 women between 30 and 70 years of age) in which samples were obtained to genotype P. gingivalis via PCR. Of 380 periodontally healthy patients, P. gingivalis was identified in 138 (36.3%), while of 139 patients with periodontitis it was identified in 121 (87.1%). In periodontally healthy subjects, FimA type I was observed in 76.1%, type II in 9.4%, type III in 7.2%, type IV in 6.5%, and type V in 29.7%; while in subjects with periodontitis, FimA type I was observed in 6.7%, type II in 66.1%, type III in 5.8%, type IV in 28.9%, and type V in 17.4%. From this, we can conclude that in periodontally healthy patients types I and V are most prevalent (p<0.00001 and p<0.0001, respectively), while types II and IV were most prevalent in patients with periodontitis (p<0.00001).
In 2002, Nakagawa et al.9 cloned a new FimA genotype, which was designated FimA Ib, indicating that it is a variant of the FimA gene I, given that the 97.1% homology between both genotypes. The researchers used a sample of 380 periodontally and systemically healthy patients (men and women aged between 30 and 70 years) and 192 patients with periodontal disease and without systemic involvement (men and women aged between 30 and 70 years). Additionally, they analyzed samples of 44 adult patients with trisomy 21 (men and women aged between 20 and 35 years) and 39 adults with mental retardation (men and women aged between 20 and 35 years). With the samples of these last patients genotipification was done with the six genotypes described and it was found that in patients with periodontitis the type II (47.4%), type Ib (13.5%), and type IV (12.5%) genotypes are relatively more prevalent compared to healthy patients, who presented frequencies of 50.4% in type I and 12.2% in type V. Likewise, this study reported that the odds ratio (OR) for the adult population in Japan (indicating the relationship between periodontitis and FimA) was higher for FimA II (OR 77.80 with 95% confidence interval of 31.0 - 195.4).
Several studies have reported the relationship between periodontal disease and cardiovascular disease, while at the same time the presence of P. gingivalis was found was found in specimens of atheromatous plaques. Nakano et al.1 demonstrated the presence of P. gingivalis bacterial DNA in 35 cardiovascular specimens collected during surgery of 35 heart valves. In 11.4% P. gingivalis DNA was detected, while of 27 specimens of atheromatous plaques, P. gingivalis DNA was detected in 7.4%. However, the same authors posed the objective of confirming the prevalence and distribution of FimA genotypes in these specimens for which they collected cardiovascular specimens during 2.5 years (between December 2004 and May 2007). These specimens consisted of 112 excised heart valves, of which 14 were through diagnosis of infective endocarditis and 98 through non-infective endocarditis, in addition to 80 specimens of atheromatous plaques with diagnosis of abdominal or thoracic aneurism, for a total of 192 specimens. From 54 of these patients who were subjected to surgery samples were taken of bacterial plaque. The results revealed that of 192 cardiovascular specimens, 20 were positive for P.gingivalis and of 56 samples of bacterial plaque, 28 were positive for P. gingivalis. In the cardiovascular samples the most frequent genotypes were FimA II (30% -6 specimens) and FimA IV (45% - 9 specimens) and of the samples of plaque higher prevalence was found in genotypes II (35.7%), I (28.6%), and IV (21.4%). Finally, the authors concluded that the FimA genotypes that have been associated to periodontitis are also frequently found in cardiovascular specimens, which suggests the possible role of the type II and IV clones at the start and progression of cardiovascular disease; although due to the low amount of cardiovascular samples positive for P. gingivalis more studies are required to confirm this hypothesis.
In 2004, in an experimental study conducted by Nakano et al. cited by Hajishengallis in 200922, it was found that FimA A II and IV genotypes were more aggressive and generated greater damage to the tissue than FimA I genotypes in a model of systemic infection disseminated in mice.
Other authors in Japan have also studied the prevalence and distribution of FimA genotypes. Miura et al.23 in 2005, analyzed the prevalence of FimA genotypes in patients with aggressive periodontitis. In that study, they included 18 patients with aggressive periodontitis and 22 periodontally healthy patients. In the patients with aggressive periodontitis, the most prevalent genotype was FimA II (40.5%), followed by FimA Ib (23.8%), and FimA I (23%). The only genotype found in healthy patients was the FimA I genotype with 100% frequency. These results are different from those reported in other studies in which the FimA IV genotype is the second most prevalent and the FimA I genotype is characteristic of healthy patients; however, in this research the genotipification was done specifically on patients with aggressive periodontitis and a variation can exist in P. gingivalis genotypes with respect to the other types of periodontitis, which could change the course and evolution of the pathology.
Guo et al. 200524 in China, investigated the distribution of P. gingivalis FimA genotypes in samples of subgingival plaque from 101 patients with chronic periodontitis. The distribution and prevalence of FimA genotypes was conducted via PCR with specific primers for each. P. gingivalis was found in 88.1% of the samples and genotype distribution was as follows: type I, 24.7%; type II, 43.8%; type III, 15.7%; type IV, 40.4%; and type V, 3.4%. It is concluded that P. gingivalis was positive in most of the subgingival samples of the patients with chronic periodontitis, and the FimA II and FimA IV genotypes were the most predominant in these patients; hence, these are the genotypes that are most associated to the development of periodontitis.
Wu et al. in 200625 in the School of Stomatology at the University of Sichuan in west China, analyzed the distribution of Porphyromonas gingivalis FimA genotypes in Chinese patients with periodontitis and their relationship between FimA genotypes and chronic periodontitis. Subgingival samples were collected from 101 patients with periodontitis. P. gingivalis was detected in 89 patients (88.1%) and the most frequent genotype was FimA II (43.8%) and FimA IV (40.4%). These two genotypes were also most prevalent in patients with moderate and severe periodontitis. The authors concluded that FimA II and FimA IV genotypes are the most prevalent in patients with periodontitis in China and these genotypes are involved in the progression of periodontal destruction.
Zhao et al. in 200726, analyzed the prevalence of FimA genotypes in 115 adult patients with chronic periodontitis in China, (67 men and 48 women between 25 and 75 years of age) and 136 periodontally healthy adult patients (44 men and 92 women between 25 and 75 years of age). Thirty of 136 healthy patients (22.1%) and 94 of 115 patients with periodontitis (85.1%) were positive for P. gingivalis. In addition, the authors found that in healthy patients 62.7% had the FimA I genotype, 16.7% FimA V genotype, and 10% FimA II genotype I; while in patients with chronic periodontitis the most prevalent genotypes were FimA II (43.6%), followed by FimA IV (30.9%) and FimA Ib (20.2%). Likewise, the study correlated the prevalence of genotypes with pocket depth, so that the most prevalent genotypes in pockets bigger or equal to 7 mm were FimA II (55.3%) and FimA IV (30.9%), while the FimA V genotype was the least prevalent in deep pockets (4.3%). Also correlated was the prevalence of FimA with bleeding upon probing, with FimA II (45.8%) and FimA IV (32.5%) genotypes being the most prevalent, and the least prevalent being the FimA V genotype (3.6%). Finally, this study also analyzed the relationship between P. gingivalis FimA genotypes and their association to other bacteria like Tannerella forsythia and Aggregatibacter actinomycetemcomitans; so that of 30 healthy patients positive for P. gingivalis, six were positive for T. forsythia and one for A. actinomycetemcomitans, while of 94 patients with chronic periodontitis positive for P. gingivalis, 60 were positive for T. forsythia and 34 for A. actinomycetemcomitans. Upon correlating the genotypes, the authors found that the frequency of coexistence of T. forsythia with FimA II was of 53.3% and the frequency of coexistence of FimA II with A. actinomycetemcomitans was 44.1%, with this genotype being the most frequently associated to other bacterial species that colonize the periodontium. Similarly, this study reported that the OR for the Chinese population regarding the relationship between the P. gingivalis FimA genotype and periodontitis was: for type I (0.97), type Ib (13.26), type II (36.62), type III (4.57), type IV (22.86), and type V (1.19), being the highest for the FimA II type, as in Japan.
In Germany, Beikler et al. 200327, conducted a study on the prevalence of FimA genotypes in 102 Caucasian patients (73 women and 29 men). The researchers initially took a universe of 203 patients of which 102 were positive for P. gingivalis (50.25%) and in whom the study was finally conducted. The research reported that 26 patients (25.5%) had FimA I, 40 patients (38.2%) had FimA II, 5 patients (4.9%) had FimA III, 19 patients (18.6%) had FimA IV and 4 patients (3.9%) had FimA V genotype. According to the authors, the differences with the results in Japan are attributed to the adaptation of the genotypes to environmental changes.
Also, Enersen et al. in 200816, carried out a study of FimA prevalence in 38 samples of patients with periodontitis from various countries like Japan, the United States, Indonesia, Canada, Switzerland, Norway, Sweden, Kenya, Germany, and France; 28 isolates from Finland and 13 isolates from Holland. The distribution of the genotypes was FimA II (34.1%), FimA IV (20.7%), FimA III (9.8%), FimA Ib (4.9%), and FimA I (3.7%). The FimA V genotype was only detected in one isolate.
For Latin America, few studies have been reported. In Brazil, Misailidis et al. 200429, analyzed 102 patients (men and women between 14 and 75 years of age, from a multiethnic Brazilian population with different periodontal conditions (healthy patients, patients with gingivitis, and patients with chronic and aggressive periodontitis). Initially, the presence of P. gingivalis was confirmed in the samples, which was positive in 2 of 25 healthy patients, 6 of 20 patients with gingivitis, 38 of 42 patients with chronic periodontitis, and 13 of 15 patients with aggressive periodontitis. The genotype distribution for this population was for patients with periodontitis; the most prevalent genotype was FimA II with 39.3%, followed by FimA Ib with 24.4%. In healthy patients, only one sample was positive for FimA II and in patients with gingivitis only two samples were positive for FimA IV. These results contrast with those reported in Japan and China, where, after FimA II the most prevalent is FimA IV, according to the authors this could be due to the racial heterogeneity of the Brazilian population and to the difference of its origins, confirming the variability of the population. This study mentions that a non-specific FimA genotype was found in 17% of the subjects studied, different from the 5.1% reported by researchers in Japan, which suggests the existence of other genotypes that have not been studied. It is concluded that the most prevalent genotype in patients with periodontitis is FimA II and that it must be considered as the most virulent and pathogenic, given that it has the greatest capacity for adhesion and invasion of tissues and that the majority of smokers had periodontitis and were positive for P. gingivalis.
Texeira et al. in 200912, conducted a study about the distribution of FimA genotypes in Brazilian patients who smoked and had chronic periodontitis, which quantitatively evaluated the levels of FimA II and FimA IV genotypes and in patients who smoked and had chronic periodontitis via reverse transcription polymerase chain reaction (RT- PCR). They found that FimA II was positive in 18 of 20 subjects and that FimA IV was found in the 20 subjects. Samples were collected from nine sites from each patient, for a total of 180 sites of which 90.5%, i.e., 152 sites were positive for P. gingivalis, of these 28% of the sites had FimA II; in contrast with FimA IV that was found in 69.6% of the sites. Also analyzed was the frequency of FimA II and FimA IV genotypes in different categories of pocket depth, observing that in this population the FimA IV genotype is more frequent in the deepest periodontal pockets. These results suggest that FimA IV is related to deeper pockets in patients with chronic periodontitis and that it is the most prevalent genotype in Brazilian patients who smoked and had periodontitis, suggesting that in this population it is the most virulent genotype.
Davila et al. in 200730, analyzed the distribution of the P. gingivalis FimA gene in patients with Diabetes type II and with periodontitis in Mexico. A total of 75 patients were selected, divided into three groups; group 1 comprised 25 healthy, non-diabetic, patients (8 men and 17 women aged between 30 and 62 years); group 2 comprised non-diabetic patients with chronic periodontitis (5 men, 20 women aged between 30 and 70 years); group 3 comprised patients with diabetes mellitus type II and with chronic periodontitis (13 men, 12 women, aged between 37 and 68 years). Subgingival samples were taken with sterile Gracey curettes, prior removal of supragingival plaque, from the distal-lingual surface of the mandibular left lateral in all the patients. DNA extraction and genotipification were carried out via PCR, using specific primers for each FimA genotype. In group 3, five patients were positive for P. gingivalis as of gene 16S rRNA, but negative for the specific primers of the FimA genotypes. These products were amplified with primers for P. gingivalis 16S rRNA and cloned in a vector (pGEM T-Easy vector®) and delivered for analysis in the Research Institute in Irapuato, Mexico (CINVESTAV-IPN). P. gingivalis was positive in group 1 (38%), group 2 (70%), and group 3 (70%). These five clones not amplified with FimA primers but with the P. gingivalis 16S rRNA universal primer had a close phylogenetic relationship with the FimA I genotype, which was analyzed and observed through a distance matrix. In all the groups, the most prevalent genotypes were type I, Ib, and II, with differences in distribution in the different groups. In group 1, the most prevalent genotype was FimA I (40%); individually and in association with other genotypes, the most prevalent was also FimA I; in group 2, FimA Ib was observed in greater proportion (20%); individually and in association it was found in 36%; in group 3, the genotypes individually found most often were I (20%) and III (20%).
A difference is noted with the high prevalence of FimA I and III genotypes in patients with diabetes type II and periodontitis. In studies in Brazil and Japan the genotype most associated to periodontitis is type II, but periodontal disease in diabetics can also be attributed to other factors that can be altered in the innate immunity, alterations in microvasculature and final metabolite glycolysis.
In Chile in 2009, Abusleme et al. genotyped 31 P. gingivalis isolates obtained from a group of 10 Chilean patients, (seven with chronic periodontitis and three with aggressive periodontitis), these isolates mostly presented the FimA II genotype (48.3%), followed by FimA I (32.2%). The lowest percentages were the FimA III (16.1%) and FimA IV (3.2%) genotypes; additionally, the study reported the presence of more than one type of Fimbriae in some patients.
In Colombia in 2009, Pérez et al.31 conducted an interesting study on FimA genotypes. They selected 15 patients, nine with chronic periodontitis and six with aggressive periodontitis who presented pockets > 7 mm and were candidates for periodontal surgery. Samples of subgingival plaques were taken prior to surgery along with a blood sample during the bacteremia after the scaling and root planing. Four blood samples were taken per patient; the first before the procedure; the second immediately at the end; the third, 15 minutes after; and the fourth, 30 minutes later. The surgical procedure was performed during 10 minutes (10 sites per patient, one minute per site). The technique used for genotipification was PCR. Six patients resulted positive via bacteremia for P. gingivalis; this finding confirms this pathogen's capacity to enter the circulatory system.
In 30 subgingival-plaque isolates a higher prevalence of FimA II was found, followed by FimA Ib, FimA III, and FimA IV. In blood isolates the most prevalent genotypes were FimA II followed by IV types, Ib and III. The most prevalent genotypes in these patients, both in the sample from subgingival plaque and strains found in blood, was FimA II, which agrees with that found by Enersen et al. in 200816, Van der Ploeg et al., in 200427, Beikler et al., 200326, Misailidis et al., in 200428, Amano et al., 200020, and Zhao et al., in 200725. In general, the results from these studies have shown that the FimA gene II is the most frequent in patients with periodontitis followed by FimA IV, while in healthy patients the most frequently found is the FimA genes I and III; however, certain differences have been found among the different populations and this has been attributed to ethnic differences existing in populations like Brazil12,29, Mexico30, and Chile.
The FimA I genotypes are less aggressive; they are associated to the first stages of the infection like colonization, invasion, and subversion of the immune response, besides lacking a capsule, which makes them less virulent bacterial strains with low invasion capacity in tissues; in contrast, FimA II and FimA IV genotypes are considered pro-inflammatory genotypes, which exhibit a more aggressive phenotype with capacity to cause damage to the tissue; generally, these genotypes are encapsulated and this gives them an advantage in terms of invasion, survival within the host, and resistance, which is related to the chronicity of the infection7,21. It has been found, in experimental studies that FimA II can adhere to epithelial cells and invade the cell more efficiently than the other genotypes and it does it through specific receptors in the host, including Integrin ∞5ß114,15.
Biological distances of porphyromonas gingivalis FimA ii and FimA iv genotypes
Various studies reported differences in the distribution of the Porphyromonas gingivalis FimA genotypes26,28 (Table 2), which were attributed to environmental factors, racial heterogeneity of the population, and systemic diseases like diabetes that alter the immune system.
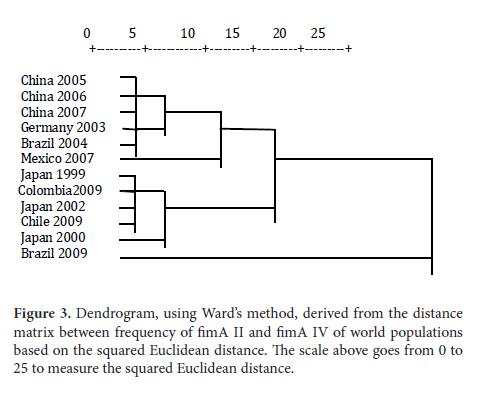
Due to this, this report used the SPSS software ver. 17.0 in English to create a distance matrix (Table 2) based on the frequency similarity of FimA II and FimA IV in several world populations from the classification of hierarchical clusters through the squared Euclidean distance. In addition, the respective dendrogram was obtained (Figure 3) by using Ward's method32 in which the elements to be related are clustered hierarchically, according to their similarity to evidence the levels of proximity among the populations included in the matrix.
The dendrogram shows the populations considered in the studies in China 200523, China 200625, and China 200726 are part of the first cluster from the frequency of FimA II and FimA IV, which indicates that the distribution of these genotypes is similar in these studies. Germany 200327 and Brazil 200429 make up the second cluster; Mexico 200730 is part of the third cluster; Japan 19997, Colombia 200931, Japan 20029, and Chile 2009 comprise the fourth cluster; Japan 200020 the fifth and Brazil 200912 the sixth cluster. These groupings into clusters show the affinity in the results of the studies conducted in the countries that are included in each of the six previously mentioned.
This rather heterogeneous distribution of the populations, bearing in mind their origin and geographic location, can be associated to the number of samples, for example, the 19997 study by Amano et al., took 93 patients with chronic periodontitis, which differs from a later study by Amano et al., in 200020, which took 380 healthy patients and 139 patients with periodontitis; this is also contrasted with studies in China, where, while Guo et al., in 200524 took a sample of 101 patients with periodontitis, Zhao et al., in 200726 worked with 115 patients with periodontitis and 136 healthy patients. This also differed with the study in Germany by Beikler et al., in 200327, who used a sample of 203 patients. In Brazil, both studies reported also present big differences in their sample number, Missailidis et al., in 200429 worked with 102 patients with periodontitis, while Texeira et al., in 200812 took a sample of 20 smokers with periodontitis.
In Mexico, Davila et al., 200730 worked with a sample of 50 patients, while in Chile, Abusleme et al., in 2009 developed a study with 10 patients with periodontitis, seven with chronic periodontitis, and three with aggressive periodontitis. In Colombia, the study conducted in 2009 by Perez et al.,31 was carried out with 15 patients, nine with chronic periodontitis, and six with aggressive periodontitis.In our country, studies are needed to permit identifying the most frequent Porphyromonas gingivalis FimA genotypes in healthy patients and in patients with different stages of periodontal disease with simple sizes calculated in such a manner that the results can be extrapolated to the Colombian population. Currently, the Periodontal Medicine Research Group of the School of Dentistry at Universidad del Valle is underway with a study which seeks to conduct genotipification of the Porphyromonas gingivalis FimA gene in healthy patients, with gingivitis and periodontitis from the city of Cali.
Clinical importance in periodontal disease
Porphyromonas gingivalis is one of the main microorganisms implied in chronic and aggressive periodontitis. In Colombia, according to a multicentric study by Lafaurie et al., in 200733, prevalence of 71.5% was found of the microorganism in a sample of 325 patients with chronic periodontitis and 14.5% in a sample of 137 healthy patients and with gingivitis. The importance of identifying those more aggressive Porphyromonas gingivalis genotypes can lead in the future to developing new therapeutic approaches aimed at eliminating this pathogen, considered one of the main etiological factors of periodontal disease. Bearing in mind that this microorganism has a good ability to invade epithelial cells and fibroblasts, in vitro34,35 the conventional scaling and planing mechanical therapy does not guarantee its total eradication; hence, it is recommended to complement this therapy with systemic antibiotics. However, in our environment the high intake of antibiotics and self-medication lead to high resistance to antibiotics, which influences on the result of these types of therapies36. Thereby, we may think of alternatives like immunoprophylaxis or immunotherapy by using target proteins important for the virulence of the microorganism, like type II and IV Fimbriline, considering these two as the most aggressive according to the results found in the studies previously referenced.
Conclusions and recommendations
Bearing in mind that Porphyromonas gingivalis is one of the main microorganisms implied in chronic and aggressive periodontitis, and considering Fimbriae as one of its main virulence factors, it is important to recognize the different genotypes of the FimA gene and their distribution and prevalence in the world population. In the different studies reported in literature a distribution may be noted, where the FimA II genotype is the most frequently found in patients with periodontitis, while in healthy patients the most frequent genotype is FimA I. This could explain the existence of strains more virulent than others and because of this some patients can be positive for P. gingivalis and not develop periodontal disease. Few studies on Porphyromonas gingivalis FimA genotypes can be related to systemic complications attributed to the spread of bacteria via bacteremia.
We recommend conducting studies in Colombia to determine the frequency and distribution of the Porphyromonas gingivalis FimA genotypes in this population to determine the most virulent strains of such to carry out primary and secondary prevention and in the future perform research that uses target proteins like FimBriline for immunoprophylaxis or inmunotherapies that lead to eliminating this pathogen from subgingival microbiota.
Conflict of interestThe author manifests having no conflict of interest regarding this study.
References
1. Nakano K, Inaba H, Nomura R, Nemoto H, Takeuchi H, Yoshioka H, et al., Distribution of Porphyromonas gingivalis FimA genotypes in cardiovascular specimens from Japanese patients. Oral Microbiol Immunol. 2008; 23: 170-2. [ Links ]
2. Armitage G. Periodontal diagnoses and classification of periodontal diseases. Periodontol. 2000; 34:9-21. [ Links ]
3. Socransky S, Hafajee A. Dental biofilms: difficult therapeutic targets. Periodontol. 2000; 28: 12-55. [ Links ]
4. Sbordone L, Bortolaia C. Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease. Clin Oral Investig. 2003; 7: 181-88. [ Links ]
5. Marcotte H, Lavoie MC. Oral microbial ecology and the role of salivary immunoglobulin A. Microbiol Mol Biol. 1998; 62: 71-109. [ Links ]
6. Holt SC, Kesavalu L, Walker S, Genco CA. Virulence factors of Porphyromonas gingivalis. Periodontol 2000; 20: 168-238. [ Links ]
7. Amano A, Nakagawa I, Kataoka K, Morisaki I, Amada S. Distribution of Porphyromonas gingivalis Strains with FimA Genotypes in Periodontitis Patients. J Clin Microbiol. 1999; 37: 1426-30. [ Links ]
8. Inaba H, Nakano K, Kato T, Nomura R, Kawai S, Kuboniwa M, et al., Heterogenic virulence and related factors among clinical isolated of Porphyromonas gingivalis with tipe II Fimbriae. Oral Microbiol Inmunol. 2008; 23: 29-35. [ Links ]
9. Nakagawa I, Amano A, Kuboniwa M, Nakamura T, Kawabata S, Hamada S. Functional differences among FimA variants of Porphyromonas gingivalis and their effects on adhesion to and invasion of human epitelial cells. Infect Immun. 2002; 70: 277-85. [ Links ]
10. Pérez-Chaparro P, Gracieux P, Lafaurie G, Donnio P, Bonnaure-Mallet M. Genotipic characterization of Porphyromonas gingivalis isolated from subgingival plaque and blood sample in positive subjects with periodontitis. J Clin Periodontol. 2008; 35: 748-53. [ Links ]
11. Takahashi Y, Davey M, Yumoto H, Gibson F, Sttardo C. Fimbria-dependent activation of pro-inflamatory molecules in Porphyromonas gingivalis infected human aortic endotelial cells. Cellul Microbiol. 2006; 8: 738-57. [ Links ]
12. Teixeira S. Mattarazo F, Feres M, Figueiredo L, de Faveri M, Simionato M, et al., Quantification of Porphyromonas gingivalis and FimA genotypes in smoker chronic periodontitis. J Clin Periodontol. 2009; 36: 482-87. [ Links ]
13. Amano A, Fujiwara T, Nagatal H, Kuboniwa M, Sharma A, Sojar H, et al., Porphyromonas gingivalis Fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dental Res. 1997; 76: 852-57. [ Links ]
14. Amano A, Nakagawa I, Okahashi N, Hamada N. Variations of Porphyromonas gingivalis Fimbriae in relation to microbial patogénesis. J Periodontal Res. 2004; 39: 139-42. [ Links ]
15. Nishiyama S, Murakami Y, Nagata H, Shizukuishi S, Kawagishi I, Yoshimura S. Involvement of minor components associated with the FimA Fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiol 2007; 153: 1916-25. [ Links ]
16. Enersen M, Olsen I, Kvalheim O, Caugant D. FimA Genotypes and Multilocus Sequence types of Porphyromonas gingivalis from patients with periodontitis. J Clin Microbiol. 2008; 46: 31-42. [ Links ]
17. Lee JY, Sojar H, Amano A, Genco R. Purification of major Fimbrial proteins of Porphyromonas gingivalis. Protein Express Purificat. 1995; 6: 496-500. [ Links ]
18. Nagano K, Hasegawa Y, Murakami Y, Nishiyama S, Yoshimura F. FimB regulates FimA Fimbriation in Porphyromonas gingivalis. J Dental Res. 2010; 89: 902-8. [ Links ]
19. Abusleme L, Pozo P, Silva N. Genotipificación de Porphyromonas gingivalis en pacientes con periodontitis. Rev Clín Period Implantol Rehabil Oral. 2009; 2: 54-8. [ Links ]
20. Amano A, Kimura R, Nakamura T, Kawabata S, Hamada S. Distribution and molecular characterization of Porphyromonas gingivalis carrying a new type of FimA gene. J Clin Microbiol. 2000; 38: 1909-14. [ Links ]
21. Amano A, Kuboniwa I, Nakagawa, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis FimA and periodontal health status. J Dental Res. 2000; 79: 1664-8. [ Links ]
22. Hajishengallis G. Porphyromonas gingivalis host interactions: open war or intelligent guerilla tactics?. Microb Infect. 2009; 11: 637-45. [ Links ]
23. Miura M, Hamachi T, Fujise O, Maeda K. The prevalence and pathogenic differences of Porphyromonas gingivalis FimA genotypes in patients whith aggressive periodontitis. J Period Res. 2005; 40: 147-52. [ Links ]
24. Guo YH, Wu YF, Liu TJ, Xiao XR, Zhou B, Zhou XP. The distribution of FimA genotype of Porphyromonas gingivalis in chronic periodontitis patients. Hua Xi Kou Qiang Yi Xue Za Zhi. 2005; 33:99-102. [ Links ]
25. Wu YF, Guo YH, Liu TJ, Xiao XR, Zhao L, Meng S, Wu AH. Distribution of FimA genotype of Porphyromonas gingivalis in Chinese periodontitis patients and its relationship with chronic periodontitis. Sichuan Da Xue Xue Bao Yi Xue Ban, 2006; 37: 101-108. [ Links ]
26. Zhao L, Wu YF, Meng S, Yang H, OuYang YL, Zhou X-D. Prevalence of FimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J Period Res. 2007; 42: 511-17. [ Links ]
27. Beikler T, Peters U, Prajaneh S, Prior K, Ehmke B, Flemmig TF. Prevalence of Porphyromonas gingivalis FimA genotypes in Caucasians. European J Oral Science. 2003; 111: 390-4. [ Links ]
28. van der Ploeg J, Giertsen E, Ludin B, Morgeli C, Zinkernagel A, Gmur R. Quantitative detection of Porphyromonas gingivalis FimA genotypes in dental plaque. FEMS Microbiol Letters. 2004; 232: 31-7. [ Links ]
29. Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MPA. Distribution of FimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microbiol Immunol. 2004; 19: 22-9. [ Links ]
30. Davila-Perez C, Amano A, Alpuche-Solis AG, Patinño-Marin N, Pontigo-Loyola AP Hamada S, et al., Distribution of genotypes of Porphyromonas gingivalis in type 2 diabetic patients with periodontitis in Mexico. J Clin Periodontol. 2007; 34: 25-30. [ Links ]
31. Pérez P, Lafaurie G, Gracieux P1, Meuric V1 Tamanai Z, Castellanos J, Bonnaure-Mallet M. Distribution of Porphyromonas gingivalis FimA genotypes in isolates from subgingival plaque and blood sample during bacteremia. Biomédica. 2009; 29: 298-306. [ Links ]
32. Ferran M. SPSS para Windows: Programación y análisis estadístico. Primera edición. Madrid. Mc Graw Hill; 2001. [ Links ]
33. Lafaurie G, Contreras A, Barón A, Botero J, Mayorga I, Jaramillo A, et al., Demographic, clinical, and microbial aspects of chronic and aggressive periodontitis in Colombia: A multicenter study. J Periodontol, 2007; 78: 629-39. [ Links ]
34. Yilmaz O, The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiol. 2008; 154: 2897-903. [ Links ]
35. Wang M, Hajishengallis G. Lipid raft dependent uptake, signalling and intracellular fate of Porphyromonas gingivalis in mouse macrophages. Cellul Microbiol. 2008; 10: 2029-42. [ Links ]
36. Jaramillo A, Betancourth M, Mayorga I, Castillo D, Aya M, Lafaurie G, et al., Perfiles antimicrobianos de bacterias subgingivales en pacientes con periodontitis en Colombia. Rev Clín Period Implantol Rehabil Oral. 2008; 1: 61-65. [ Links ]