Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Colombia Médica
On-line version ISSN 1657-9534
Colomb. Med. vol.44 no.2 Cali Apr./Jun. 2013
Review Point
Malaria vaccines: high-throughput tools for antigens discovery with potential for their development
Vacunas contra la malaria: estrategias de alto rendimiento para el descubrimiento de antígenos con potencial para su desarrollo
Nora Céspedesa, Andrés Vallejoa, Myriam Arévalo-Herrerab, Sócrates Herrerac
a International Vaccine Center. E-mail: ncespedes@inmuno.org, avallejo@inmuno.org
b Universidad del Valle. E-mail: marevalo@inmuno.org
c Corresponding author, E-mail: sherrera@inmuno.org
Article history: Received: Sep 3, 2012; Received in revised form: Sep 20, 2012; Accepted: Jan 15, 2013.
Abstract
Malaria is a disease induced by parasites of the Plasmodium genus, which are transmitted by Anopheles mosquitoes and represents a great socio-economic burden Worldwide. Plasmodium vivax is the second species of malaria Worldwide, but it is the most prevalent in Latin America and other regions of the planet. It is currently considered that vaccines represent a cost-effective strategy for controlling transmissible diseases and could complement other malaria control measures; however, the chemical and immunological complexity of the parasite has hindered development of effective vaccines. Recent availability of several genomes of Plasmodium species, as well as bioinformatic tools are allowing the selection of large numbers of proteins and analysis of their immune potential. Herein, we review recently developed strategies for discovery of novel antigens with potential for malaria vaccine development.
Keywords: Malaria, vaccines, Plasmodium, Antigens, peptides, Recombinant Proteins.
Resumen
La malaria es una de las enfermedades transmisibles de mayor impacto socio-económico a escala mundial, y es inducida por parásitos del género Plasmodium transmitidos por mosquitos del genero Anopheles. El Plasmodium vivax ocupa el segundo lugar en prevalencia mundial, pero es la especie más frecuente en América Latina y otras regiones del planeta. Se considera que las vacunas representan una estrategia costo-efectiva para el control de enfermedades transmisibles y que podrían complementar las demás medidas de control de la malaria; sin embargo, la complejidad química e inmunológica del parasito han dificultado el desarrollo de vacunas efectivas. La reciente accesibilidad a los genomas de varias especies de Plasmodium, y el desarrollo de herramientas bioinformáticas están permitiendo la selección de numerosas proteínas y el análisis de su potencial inmunológico. Aquí revisamos las estrategias recientes para el descubrimiento de nuevos antígenos para el desarrollo vacunas contra la malaria.
Palabras clave: Malaria, vacunas, Plasmodium, Antígenos, péptidos, proteínas recombinantes.
Epidemiological importance of malaria
Malaria represents a global public health problem that hinders socio-economic development in vast regions of the world, particularly of the planet´s Tropical and sub-Tropical areas. It is calculated that ∼3-billion people from over 100 countries are exposed to the infection by one or more Plasmodium1 species and it is estimated that in 2010 there were over 216-million clinical cases, over 650.000 of which were lethal. P. falciparum is the most abundant and virulent species, followed by P. vivax , which although producing lower mortality causes incapacitating and recurrent disease2. Plasmodium. vivax , coexists with P. falciparum in vast zones of the planet and it is prevalent in regions of Asia, Oceania, and Latin America where it is estimated to produce between 70 and 80-million clinical cases each year2. Mortality produced by malaria is higher in Africa, mainly in children younger than five years of age and in pregnant women infected by P. falciparum and although mortality is present to a lesser degree in infections by P. vivax, a significant number of lethal cases has been recently documented in high-transmission regions like India and Brazil3,4.
Strategies and global programs of malaria control
Although the classical measures of malaria control, like early diagnosis, timely and efficient treatment, and mosquito control have contributed significantly to reducing the malaria distribution map5, it is currently considered that the development and possible application of malaria vaccines would contribute significantly and cost-effectively to reduce the impact of malaria in zones affected by the disease, and would favor its elimination in zones that currently have lower transmission. During the last decade, several global initiatives aimed at efficient malaria control have been developed including: the program denominated Roll Back Malaria (RBM), the Malaria Elimination Group (MEG), and, more recently, the Malaria Eradication Research Agenda (malERA). Additionally, the Global Fund (GF) has been, since 2002, perhaps the main funding source for malaria control Worldwide.
Until now, no malaria vaccine has been licensed for massive application in populations; however, rising evidence indicates the feasibility of developing vaccines. Firstly, in individuals exposed to malarial infection in endemic areas, the naturally acquired immunity accumulates progressively during the first two decades of life and results in decreased clinical severity of the disease and mortality6. Secondly, experimental immunization of non-immune volunteers with sporozoites previously attenuated through irradiation has demonstrated that up to 90% of the individuals vaccinated develop sterile protection against the experimental infection7. Thirdly, it has been shown that passive transfer of immunoglobulin from immune adults to naive volunteers eliminates the circulating parasites8. Additionally, it has been recently shown that it is possible to protect endemic communities from P. falciparum, at least partially, through immunization with an experimental vaccine, the RTS,S based on the P. falciparum CS protein9.
Importance of vaccines as control strategy
At least three levels have been contemplated of the Plasmodium cycle in which the parasite would be most susceptible to the immunological attack induced by a vaccine: the pre-erythrocytic stage (sporozoites and liver stages) and the asexual erythrocytic phase based on its capacity to stimulate humoral and cellular immune response. During the pre-erythrocytic stage, antibodies can inhibit invasion of sporozoites to the liver10 and, hence, prevent hepatic development of the parasite and the ulterior disease; cytokines like IFN-γ produced by T CD4+ and T CD8+ cells would contribute to halt intracellular development of hepatic schizonts11. During the erythrocytic stage, the presence of antibodies can, through different mechanisms, prevent invasion of the parasite to the erythrocytes12 and also, in red blood cells, oxygen radicals can destroy intracellular parasites13. A third level in which the parasite´s life cycle can be interrupted is the sporogonic phase, which occurs in the mosquito´s intestine. During this phase, it is possible to interrupt the fertilization process and ookinete invasion to the mosquito´s intestinal cells, preventing development of the parasite within the mosquito and, hence, its transmission to other susceptible individuals14.
Although vaccines must individually prove their efficacy, it is considered essential to focus efforts on generating formulations that include all the stages of the parasite´s cycle. Additionally, given the epidemiological distribution of malaria throughout the world, a functional vaccine against it must include components from at least the two most abundant species, P. falciparum and P. vivax. Accomplishing this aim in the near future is not easy because of the differential development of research on P. falciparum and on P. vivax. This review seeks to describe the use of high-performance tools and recent progress in identifying new antigen candidates for vaccines against P. falciparum and P. vivax during erythrocytic and pre-erythrocytic stages.
Strategies to discover antigens with potential for vaccine development:
Classical strategies Vaccine production from inactive living, attenuated, or dead organisms, which have been employed to develop several of the vaccines for use in humans is not functional for diseases like malaria due to numerous factors like: contamination of the formulation with components from human cells, loss of immunogenicity, and difficulty in logistics for their production15,16. Thereby, the approach used during the last two to three decades to design a malaria vaccine has been based on identifying "subunits" of the parasite, such as complete antigens or their fragments, which have been mainly produced as synthetic peptides and recombinant proteins derived from sporozoite, merozoite, or gametocyte stages. Additionally, other methods have been tried like the production of vaccines from DNA and recombinant viruses. Numerous antigens, particularly from P. falciparum have been produced and analyzed in preclinical studies (on animals) in which their immunogenicity and lack of toxicity have been determined; essential conditions for their advance to the clinical development phase in humans. This process has given way to currently most advanced proteins being tested in clinical and preclinical phases (Table 1). Particular emphasis has been made on the P. falciparum circumsporozoite (CS) protein, which has reached maximum progress in its clinical development, recently accomplishing its analysis during Phase III clinical studies. This vaccine denominated Pf-RTS,S has demonstrated the capacity to induce protection against clinical and severe malaria in African children9, and although most recent studies registered protection of ∼30%, the progress and learning accomplished during its analysis is of great value. The homologous protein in P. vivax has been analyzed in clinical trials in Phases I17 and, currently, a Phase II study is under way. Several other vaccines have reached clinical phases and are reviewed in available literature18,19.
New Strategies Of Antigen Discovery
Use of the Expressed Sequence Tags (EST) technology permitted, during 2000-2003, describing the human genome and the genomes of multiple microorganisms of biological, agricultural, archaeological, and medical interest20-22. This vertiginous development permitted learning that parasites from the Plasmodium genus have genomes composed by between 5000 and 6000 genes. Availability of genomes from P. falciparum, P. vivax and from some other species, as well as progress in bioinformatics, genomics, and proteomics, are permitting the development of high-performance methods to select, clone/synthesize, and analyze a large number of proteins of these parasites, and it is probable that it accelerates development of malaria vaccines. Specifically, proteomic analyses have indicated that throughout the parasite´s life cycle at least 5,440 proteins are expressed by P. falciparum and 5,321 by P. vivax23,24. Given this enormous multiplicity of proteins, it becomes more evident that the clinical immunity developed under natural and experimental conditions against human malaria is probably induced by multiple components of the parasite whose identification and analysis is only possible by using high-performance techniques.
Proteomics-based tools for discovery of new vaccine candidates
As a result of this important progress, the development of high-performance technologies has emerged as great promise, and terms like transcriptomics, metabolomics, lipidomics, and proteomics are increasingly more common in biomedical literature, and it is expected that in the following years we will see the huge scope of technologies25. In light of the need to analyze the big volume of information produced on only one experiment based on high-performance technology, recently numerous tools have been developed for information analysis, which currently facilitate data mining and efficient development of complex studies employing "omic" technology.
Microarrays of proteins
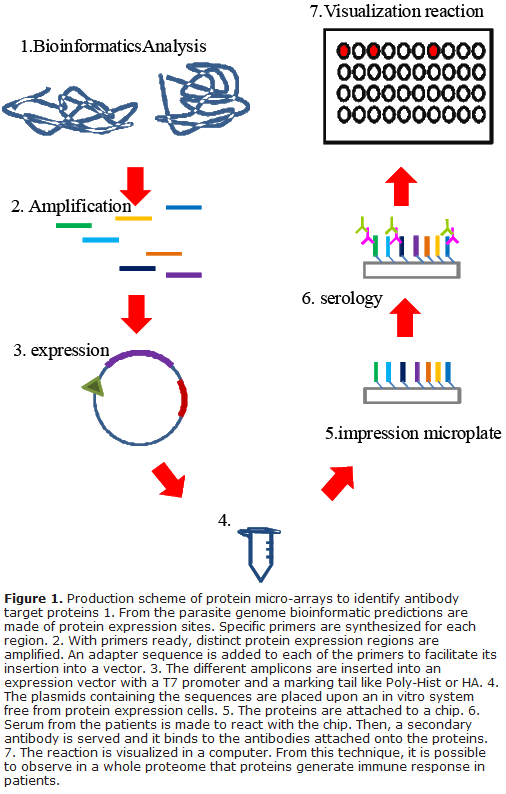
Consist of protein libraries assembled on a same format generated through cloning and expression of large fragments of the genome of microorganisms, in this case of P. falciparum and P. vivax, available to simultaneously conduct studies, for example, of reactivity with antibodies. In contrast to classical techniques that permitted analysis of individual genes and proteins, this proteomic technology permits simultaneous analysis of immunoreactive profile of hundreds of the organism´s significant proteins or of its fragments and represent one of the most attractive approaches for large-scale discovery of new vaccine candidates. One of the pioneer laboratories in malaria proteomics research is led by Dr. P. Felgner at the University of California (USA). His group developed a high-performance system for protein expression called "PCR Express", through which complete proteomes of any microorganism are generated26 (Fig 1).
Compared to conventional cloning methods in plasmid expression, transformation, and growth vectors in bacteria, "PCR express" offers various advantages for the production of transcriptionally active genes, given that these use up a large amount de time and require intensive labor, above all when seeking to simultaneously clone a large number of genes. The difference lies in that PCR express is based on a sound and practical approach for production transcriptionally active PCR fragments (TAPF) in two sequential PCR reactions. Technically, the process consists of two stages; a first stage uses gene-specific primers to amplify the gene of interest, while a second stage carries out nested PCR, which uses a mix of DNA fragments to add promoter and terminator sequences to each fragment. This makes the TAPFs equally active to super-coiled plasmid DNA, produced in in-vitro and in-vivo transfection assays and, hence, can be used as DNA vaccines27.
As in DNA vaccines and in recombinant viruses containing Plasmodium protein inserts previously described28, TAPFs can also be quickly transferred to plasmid vectors through homologous recombination, offering a high-performance cloning method that does not require using restriction enzymes or ligation reactions. The TAPF technology has been used with different pathogens to evaluate serum antibody titers from people or animals, vaccinated or infected naturally, to identify antigens recognized by the immune system after vaccination or infection with such microorganism29. Based on this technology, the most reactive antigens are selected for evaluation in different kind of studies, e.g., immunogenicity to determine their potential for vaccine development or to design immune-diagnostic methods. Until now, micro-arrays corresponding to ∼30 microorganisms have been assessed, including viruses, bacteria, and pathogen parasites (Table 2).
Progress in the discovery of new Plasmodium antigens using protein microarrays
During the Plasmodium life cycle, over 5000 proteins are expressed and it is not exactly clear which of these antigens mediate the clinical immunity and the protecive immunity observed on individuals from endemic areas or on individuals vaccinated with the parasite attenuated via irradiation7. Preliminary studies suggest that a large number of antigens are recognized30,31 by the host´s immune system, highlighting the importance of identifying the repertoire of antigens and epitopes from the different development phases of the parasite implied in induction of clinical immunity. Recently, high-performance tools have been used to identify immunodominant antigens and preliminary definition of immunoreactivity profiles among groups of individuals with different levels of immunity to malaria32. A first study selected a panel of 250 P. falciparum proteins chosen specifically to evaluate immune response in volunteers immunized with sporozoites attenuated via radiation, mainly in hepatic stages and with sporozoites, representing 4.75% of the totality of the genome, which were cloned, expressed and included in protein micro-arrays32. The reactivity of these micro-arrays was then evaluated against serum samples from individuals vaccinated with irradiated sporozoites and compared to that of serum from individuals from endemic areas of Kenya, with different degrees of exposure to malaria. In this study, it was noted that subjects naturally exposed to malaria in Kenya reacted with greater intensity to a large number of antigens than individuals vaccinated with irradiated sporozoites. Also, among the group of subjects immunized with irradiated sporozoites, those protected against infectious challenge react with greater intensity and at a higher number of antigens than those unprotected. Additionally, it was noted that 56 of a total of 72 proteins that were the most highly reactive had not been previously characterized, which could represent a potential for development of malaria vaccines jo P. falciparum.
In another study held in Mali with serum samples from 220 individuals ranging in age between 2 and 10 years and between 18 and 25 years31, it was observed that the average number of proteins recognized by individuals exposed to the infection increased with age. Furthermore, it was found that reactivity in children increases dramatically during periods of high transmission of malaria. In contrast, the number of proteins recognized by the adults does not vary significantly during these periods. Finally, these analyses provided information about the patterns of reactivity against P. falciparum proteins based on the phases of the life cycle in which the proteins are expressed, as well as their sub-cellular location and other proteomic characteristics.
For P. vivax, few genes of the ∼5500 encoded by the genome from the Salvador I (Sal I) strain23 have been assessed. Recently, 10 proteins from pre-erythrocytic stages were identified, which were widely recognized by serum from individuals from endemic regions of the Colombian Pacific Coast (n=60). These proteins were identified from a panel of 91 antigens evaluated in micro-arrays33 that are currently available for additional characterization.
Similar to the protein micro-arrays, the peptide micro-arrays are now being used for functional evaluation of proteins. Peptides are quite stable functionally, capable of maintaining their activity under most reaction conditions, which gives them advantages in application like micro-array. In general, depending on the peptide micro-array preparation method, these can be classified into: (a) in situ, or (b) synthesis followed by immobilization.
As the name implies, in situ micro-arrays are synthesized directly on the solid surface. The technique consists of dispensing a small volume of solutions containing amino acids and other coupling reagents to a designated point on a membrane.
In the second case, peptides are previously synthesized by using conventional equipment and methods and then nanoliters of peptide solutions are transferred to the solid surface. This approach is much more efficient because each peptide needs to be synthesized only once34.
Peptide micro-arrays have been used in identifying ligands or substrates of target molecules of interest, as well as in evaluating activity and enzyme and protein bonding35. Peptide micro-arrays can be used, for example, to identify epitopes and evaluate the response of the immune system to different pathogens. Using this technology, Wiley et al., demonstrated that the recognition profile of different epitopes from a P. vivax protein changes en individuals vaccinated with the antigen formulated in different adjuvants35.
Using bioinformatic tools to select structural motifs in Plasmodium proteins
In spite of the great value of massively identifying immunodominant proteins, direct identification of relevant epitopes represents a high valuable complementary strategy. Many of the epitopes recognized by antibodies represent three-dimensional surfaces of an antigen molecule that precisely interact with the bonding surfaces of the corresponding antibodies; these epitopes can be linear or conformational. Linear epitopes are formed by a continuous sequence of amino acids, while conformational epitopes depend on the position of the amino acids in the protein´s three-dimensional structure, which is determined by the combination of its alpha (α) helix, folded beta (β) sheet, or β coil structures. It has been shown that α double-helix structures contain abundant epitopes from B cells that have proven to be targets of Plasmodium growth inhibiting antibodies36 and other pathogen agents37. Additionally, it is estimated that most B epitopes are structural and that only 10% of the antibodies induced by immune response are aimed against linear epitopes38.
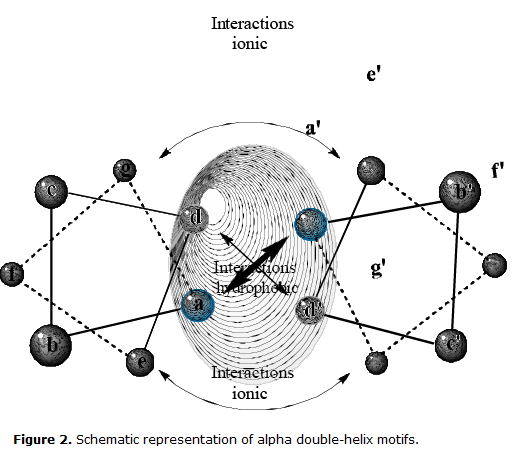
Production of protein fragments containing conformational epitopes is of great importance, but it represents a huge synthesis technical challenge39. Fragments of synthesized proteins need to acquire stable structures that mimic the native structure and, hence, can be recognized by antibodies. Further, the challenge exists of identifying such fragments in protein sequences throughout the genome/proteome. Recently, specific bioinformatic algorithms have been developed to select sequences containing α double-helix motifs. These motifs form stable structures characterized by the presence of repetitions from seven residues from amino acids (abcdefg) with hydrophobic residues located in positions a and d, and hydrophilic residues in the remaining positions (Fig 2), which are generally monomorphic and react with antibodies that are reactive with the native form of the native protein. These structures are easily identifiable with bioinformatic tools40, which significantly reduces antigen selection time.
Recent progress in chemical synthesis techniques have permitted the production of 95 fragments of 30-40 amino acids of length corresponding to P. falciparum proteins, which contained α double-helix motifs. All the antigens selected were evaluated to determine their antigenicity by using a panel of serum from donors from endemic zones41, which permitted identifying ∼70 new proteins with variable length between 200 and 10,000 amino acids. Thereafter, the immunogenicity of some of these fragments was evaluated in murine models. Functional assays conducted using specific antibodies against the different fragments, demonstrated their capacity to inhibit in vitro development of the parasite in antibody-dependent inhibition trials (ADIT), and it was also demonstrated that these structures are highly conserved42. A Phase 1 clinical trial is currently analyzing fragment P27A of 104 amino acids derived from the P. falciparum protein PFF0165c selected by using this methodology43; in addition, the immunogenicity of the Pf-P181 polypeptide, also selected through this methodology and which contains fragments from three different proteins bound by a nonimmunogenic connector (diethylene glycol), was recently evaluated during preclinical trials44.
The same technology is being currently used in identifying P. vivax antigens. A total of 52 peptides containing α double-helix motifs were recently selected from 150 proteins of P. vivax erythrocytic stages. Fragments of variable lengths between 30 and 50 residues were synthesized by using F-moc solid phase chemistry and used to determine their antigenicity by comparing their reactivity with serum from individuals naturally exposed to malaria in hyper-endemic areas of Papua New Guinea (PNG) (District of Maprik) and from areas of medium and low transmission in Colombia (Tumaco and Tierralta). In general, higher reactivity was observed when using serum from individuals from PNG; a total of 10 fragments have been preselected because of their high reactivity with serum from PNG and from Colombia and they are being used in immunogenicity assays in mice.
Conclusions
The large number of proteins produced by Plasmodium and their great diversity demand the use of high-performance tools for their identification, production, and analysis. During the last decade, important progress in bioinformatics and biotechnology has permitted the construction of micro-arrays of proteins produced via recombinant technology or as synthetic peptides, which have permitted identifying over 400 new antigens from pre-erythrocytic and erythrocytic phases with possible functions in the natural immunity acquired for P. falciparum and P. vivax. However, although this progress permits studying more efficiently the response profile of antibodies associated to immunity acquired naturally or through vaccination (antigenicity), immunogenicity analyses of proteins or epitopes selected still require in-vivo models which are not currently scalable. The important progress in the study of immune response against Plasmodium through high-performance methods now generates exceptional conditions for identifying new vaccine candidates for their clinical development.
Acknowledgments
This review and several of the studies presented were developed at the CIV with COLCIENCIAS support (Contract: 278-2008, Contract 527-2009, and Contract 360-2011) and by the United States National Institute of Allergy and Infectious Diseases (R01 grant AI05759206).
References
1. WHO. World malaria report 2011. Geneva: World Health Organization, 2011. [ Links ]
2. Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97-106. [ Links ]
3. Kochar DK, Das A, Kochar SK, Saxena V, Sirohi P, Garg S, et al. Severe Plasmodium vivax malaria: A report on serial cases from bikaner in northwestern india. Am J Trop Med Hyg. 2009;80:194-8. [ Links ]
4. Lacerda MV, Hipolito JR, Passos LN. Chronic Plasmodium vivax infection in a patient with splenomegaly and severe thrombocytopenia. Rev Soc Bras Med Trop. 2008;41:522-3. [ Links ]
5. Feachem RGA. Shrinking the malaria map: A prospectus on malaria elimination. San Francisco: The Global Health Group; 2009. [ Links ]
6. Baird JK, Masbar S, Basri H, Tirtokusumo S, Subianto B, Hoffman SL. Age-dependent susceptibility to severe disease with primary exposure to Plasmodium falciparum. J Infect Dis. 1998;178:592-5. [ Links ]
7. Clyde DF. Immunity to falciparum and vivax malaria induced by irradiated sporozoites: A review of the university of maryland studies, 1971-75. Bull World Health Organ. 1990;68 Suppl:9-12. [ Links ]
8. Sabchareon A, Burnouf T, Ouattara D, Attanath P, Bouharoun-Tayoun H, Chantavanich P, et al. Parasitologic and clinical human response to immunoglobulin administration in falciparum malaria. Am J Trop Med Hyg. 1991;45:297-308. [ Links ]
9. Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BG, Kabwende AL, et al. A phase 3 trial of rts,s/as01 malaria vaccine in african infants. N Engl J Med. 2012;367:2284-95. [ Links ]
10. Jones TR, Ballou WR, Hoffman SL. Antibodies to the circumsporozoite protein and protective immunity to malaria sporozoites. Prog Clin Parasitol. 1993;3:103-117. [ Links ]
11. Sedegah M, Sim BK, Mason C, Nutman T, Malik A, Roberts C, et al. Naturally acquired cd8+ cytotoxic t lymphocytes against the Plasmodium falciparum circumsporozoite protein. J Immunol. 1992;149:966-71. [ Links ]
12. Bouharoun-Tayoun H, Attanath P, Sabchareon A, Chongsuphajaisiddhi T, Druilhe P. Antibodies that protect humans against Plasmodium falciparum blood stages do not on their own inhibit parasite growth and invasion in vitro, but act in cooperation with monocytes. J Exp Med. 1990;172:1633-41. [ Links ]
13. Kharazmi A, Jepsen S, Andersen BJ. Generation of reactive oxygen radicals by human phagocytic cells activated by Plasmodium falciparum. Scand J Immunol. 1987;25:335-41. [ Links ]
14. Mendis KN, Munesinghe YD, de Silva YN, Keragalla I, Carter R. Malaria transmission-blocking immunity induced by natural infections of Plasmodium vivax in humans. Infect Immun. 1987;55:369-72. [ Links ]
15. Hoffman SL, Billingsley PF, James E, Richman A, Loyevsky M, Li T, et al. Development of a metabolically active, non-replicating sporozoite vaccine to prevent Plasmodium falciparum malaria. Hum Vaccin. 2010;6:97-106. [ Links ]
16. Pinder M, Moorthy VS, Akanmori BD, Genton B, Brown GV. Malvac 2009: Progress and challenges in development of whole organism malaria vaccines for endemic countries, 3-4 june 2009, dakar, senegal. Vaccine. 2010;28:4695-702. [ Links ]
17. Herrera S, Fernandez OL, Vera O, Cardenas W, Ramirez O, Palacios R, et al. Phase i safety and immunogenicity trial of Plasmodium vivax cs derived long synthetic peptides adjuvanted with montanide isa 720 or montanide isa 51. Am J Trop Med Hyg. 2011;84:12-20. [ Links ]
18. Arevalo-Herrera M, Chitnis C, Herrera S. Current status of Plasmodium vivax vaccine. Hum Vaccin. 2010;6:124-32. [ Links ]
19. Schwartz L, Brown GV, Genton B, Moorthy VS. A review of malaria vaccine clinical projects based on the who rainbow table. Malar J. 2012;11:11. [ Links ]
20. Robinson K. Emerging trends in genetic-based medical diagnostics. Clin Leadersh Manag Rev. 2005;19:E2. [ Links ]
21. Bellen HJ, Levis RW, He Y, Carlson JW, Evans-Holm M, Bae E, et al. The drosophila gene disruption project: Progress using transposons with distinctive site specificities. Genetics. 2011;188:731-43. [ Links ]
22. Qiu YL, Yu J. Azolla--a model organism for plant genomic studies. Genomics Proteomics Bioinformatics. 2003;1:15-25. [ Links ]
23. Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455:757-63. [ Links ]
24. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498-511. [ Links ]
25. Wang D, Bodovitz S. Single cell analysis: The new frontier in 'omics'. Trends Biotechnol. 2010;28:281-90. [ Links ]
26. Liang X, Teng A, Braun DM, Felgner J, Wang Y, Baker SI, et al. Transcriptionally active polymerase chain reaction (tap): High throughput gene expression using genome sequence data. J Biol Chem. 2002;277:3593-98. [ Links ]
27. Saade F, Petrovsky N. Technologies for enhanced efficacy of DNA vaccines. Expert Rev Vaccines. 2012;11:189-209. [ Links ]
28. Moorthy VS, Good MF, Hill AV. Malaria vaccine developments. Lancet. 2004;363:150-6. [ Links ]
29. Vigil A, Davies DH, Felgner PL. Defining the humoral immune response to infectious agents using high-density protein microarrays. Future Microbiol. 2010;5:241-51. [ Links ]
30. Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003;100:9952-7. [ Links ]
31. Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, et al. A prospective analysis of the ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci U S A. 2010;107:6958-63. [ Links ]
32. Doolan DL, Mu Y, Unal B, Sundaresh S, Hirst S, Valdez C, et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics. 2008;8:4680-94. [ Links ]
33. Molina DM, Finney OC, Arevalo-Herrera M, Herrera S, Felgner PL, Gardner MJ, et al. Plasmodium vivax pre-erythrocytic-stage antigen discovery: Exploiting naturally acquired humoral responses. Am J Trop Med Hyg. 2012. [ Links ]
34. Wu H, Ge J, Uttamchandani M, Yao SQ. Small molecule microarrays: The first decade and beyond. Chem Commun (Camb). 2011;47:5664-70. [ Links ]
35. Uttamchandani M, Yao SQ. The expanding world of small molecule microarrays. Methods Mol Biol. 2010;669:1-15. [ Links ]
36. Singh S, Soe S, Roussilhon C, Corradin G, Druilhe P. Plasmodium falciparum merozoite surface protein 6 displays multiple targets for naturally occurring antibodies that mediate monocyte-dependent parasite killing. Infect Immun. 2005;73:1235-8. [ Links ]
37. Tripet B, Kao DJ, Jeffers SA, Holmes KV, Hodges RS. Template-based coiled-coil antigens elicit neutralizing antibodies to the sars-coronavirus. J Struct Biol. 2006;155:176-94. [ Links ]
38. Barlow DJ, Edwards MS, Thornton JM. Continuous and discontinuous protein antigenic determinants. Nature. 1986;322:747-8. [ Links ]
39. Corradin G, Villard V, Kajava AV. Protein structure based strategies for antigen discovery and vaccine development against malaria and other pathogens. Endocr Metab Immune Disord Drug Targets. 2007;7:259-65. [ Links ]
40. Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162-4. [ Links ]
41. Villard V, Agak GW, Frank G, Jafarshad A, Servis C, Nebie I, et al. Rapid identification of malaria vaccine candidates based on alpha-helical coiled coil protein motif. PLoS One. 2007;2:e645. [ Links ]
42. Kulangara C, Kajava AV, Corradin G, Felger I. Sequence conservation in Plasmodium falciparum alpha-helical coiled coil domains proposed for vaccine development. PLoS One. 2009;4:e5419. [ Links ]
43. Olugbile S, Kulangara C, Bang G, Bertholet S, Suzarte E, Villard V, et al. Vaccine potentials of an intrinsically unstructured fragment derived from the blood stage-associated Plasmodium falciparum protein pff0165c. Infect Immun. 2009;77:5701-9. [ Links ]
44. Olugbile S, Villard V, Bertholet S, Jafarshad A, Kulangara C, Roussilhon C, et al. Malaria vaccine candidate: Design of a multivalent subunit alpha-helical coiled coil poly-epitope. Vaccine. 2011;29:7090-9. [ Links ]
45. Mason JM, Arndt KM. Coiled coil domains: Stability, specificity, and biological implications. Chembiochem : a European J Chem Biol.. 2004;5:170-6. [ Links ]
46. Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Kone AK, Guindo AB, et al. Safety and immunogenicity of an ama1 malaria vaccine in malian children: Results of a phase 1 randomized controlled trial. PLoS One. 2010;5:e9041. [ Links ]
47. Sheehy SH, Duncan CJ, Elias SC, Collins KA, Ewer KJ, Spencer AJ, et al. Phase Ia clinical evaluation of the Plasmodium falciparum blood-stage antigen msp1 in chad63 and mva vaccine vectors. Mol Ther. 2011;19:2269-76. [ Links ]
48. McCarthy JS, Marjason J, Elliott S, Fahey P, Bang G, Malkin E, et al. A phase 1 trial of MSP2-c1, a blood-stage malaria vaccine containing 2 isoforms of MSP2 formulated with montanide(r) isa 720. PLoS One. 2011;6:e24413. [ Links ]
49. Belard S, Issifou S, Hounkpatin AB, Schaumburg F, Ngoa UA, Esen M, et al. A randomized controlled phase Ib trial of the malaria vaccine candidate gmz2 in african children. PLoS One. 2011;6:e22525. [ Links ]
50. Herrera S, Bonelo A, Perlaza BL, Fernandez OL, Victoria L, Lenis AM, et al. Safety and elicitation of humoral and cellular responses in colombian malaria-naive volunteers by a Plasmodium vivax circumsporozoite protein-derived synthetic vaccine. Am J Trop Med Hyg. 2005;73:3-9. [ Links ]
51. Wu Y, Ellis RD, Shaffer D, Fontes E, Malkin EM, Mahanty S, et al. Phase 1 trial of malaria transmission blocking vaccine candidates Pfs25 and Pvs25 formulated with montanide isa 51. PLoS One. 2008;3:e2636. [ Links ]
52. Yoon D, Kim H, Suh-Kim H, Park RW, Lee K. Differentially co-expressed interacting protein pairs discriminate samples under distinct stages of hiv type 1 infection. BMC Syst Biol. 2011;5 Suppl 2:S1. [ Links ]
53. Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, et al. Profiling the humoral immune response to infection by using proteome microarrays: High-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005;102:547-52. [ Links ]
54. Luevano M, Bernard HU, Barrera-Saldana HA, Trevino V, Garcia-Carranca A, Villa L, et al. High-throughput profiling of the humoral immune responses against thirteen human papillomavirus types by proteome microarrays. Virol. 2010;405:31-40. [ Links ]
55. Kalantari-Dehaghi M, Chun S, Chentoufi AA, Pablo J, Liang L, Dasgupta G, et al. Discovery of potential diagnostic and vaccine antigens in herpes simplex virus 1 and 2 by proteome-wide antibody profiling. J Virol. 2012;86:4328-39. [ Links ]
56. Vizoso Pinto MG, Pfrepper KI, Janke T, Noelting C, Sander M, Lueking A, et al. A systematic approach for the identification of novel, serologically reactive recombinant varicella-zoster virus (vzv) antigens. Virol J. 2010;7:165. [ Links ]
57. Fernandez S, Cisney ED, Tikhonov AP, Schweitzer B, Putnak RJ, Simmons M, et al. Antibody recognition of the dengue virus proteome and implications for development of vaccines. Clin Vaccine Immunol. 2011;18:523-32. [ Links ]
58. Kunnath-Velayudhan S, Davidow AL, Wang HY, Molina DM, Huynh VT, Salamon H, et al. Proteome-scale antibody responses and outcome of mycobacterium tuberculosis infection in nonhuman primates and in tuberculosis patients. J Infect Dis. 2012;206:697-705. [ Links ]
59. Ceroni A, Sibani S, Baiker A, Pothineni VR, Bailer SM, LaBaer J, et al. Systematic analysis of the IgG antibody immune response against varicella zoster virus (vzv) using a self-assembled protein microarray. Mol Biosyst. 2010;6:1604-10. [ Links ]
60. Vieira ML, Pimenta DC, de Morais ZM, Vasconcellos SA, Nascimento AL. Proteome analysis of leptospira interrogans virulent strain. Open Microbiol J. 2009;3:69-74. [ Links ]
Céspedes N, Vallejo A, Arevalo-Herrera M, Arevalo M, Herrera S. Malaria vaccines: high-throughput tools for antigen discovery with development potential.Colomb.Med. 2013; 44 (2):121-8