Remark
| 1) Why was this study done? |
| LRBA deficiency patients exhibit a clinically heterogeneous syndrome. The main clinical complication of LRBA deficiency is immune dysregulation. Enteropathy, autoimmune hemolytic anemia and idiopathic thrombocytopenic purpura are the most common clinical manifestation. In more than half of patients are at low levels: IgM and IgG or IgA and IgG, IgA only or IgG only. To date, no patients with this condition have been reported in Colombia. Was evaluated of mutations in LRBA in patients from Colombia with predominantly antibody deficiencies such as CVID, XLA, IHG and SIgAD, autoimmune manifestations such as ITP and SLE and/or enteropathies and determine if the cause of the clinical phenotype in these patients is mutations in LRBA or are other genes responsible for the pathogenesis |
| 2) What did the researchers do and find? |
| Six (n= 112) patients showed decreased in LRBA protein expression. We observed a reduction in two patients with CVID, two with XLA and in two with IHG. These patients exhibit different phenotypic and immunologic characteristic. However, all patients presented with a reduction in IgG and recurrent infections by otitis and sinusitis, and five presented with gastropathy. We didn’t find pathogenic or potentially pathogenic variants neither in LRBA nor in other immune-related genes. |
| 3) What do these findings mean? |
| The expression of LRBA identifies a lot in the CMSP between different DS and patients. Six patients exhibited a decrease in the expression of this protein. No genes associated with defective LRBA production by these cells were detected in these patients. Homozygous mutations in LRBA cause deficiency that affects protein expression. However, reduced LRBA expression in patients without homozygous mutations or in associated genes were present. These results detect the other genetic mechanisms, epigenetic or environmental, may be regulating the expression of LRBA. |
Introduction
Primary immunodeficiency diseases (PID) are a heterogeneous group of inheritable genetic disorders that cause quantitative and/or functional alterations in different mechanisms involved in the immune response 1. PIDs are considered “rare” diseases, but most prevalence estimates have been based on selected populations (e.g. specialty clinics, disease registries)2. However, these the frequencies are much higher than the reported estimates based on the registry data suggesting both underreporting and ascertainment bias. Therefore, the data obtained from registries does not provide adequate coverage to estimate prevalence or age distribution of PID 2.
LPS-responsive beige -like anchor protein (LRBA) deficiency is a PID caused by loss of LRBA protein expression. LRBA deficiency is an autosomal recessive disorder caused by either homozygous or compound heterozygous mutations in LRBA gene 3. LRBA deficiency results in a clinically variable syndrome with a wide spectrum of clinical manifestations. The main clinical LRBA deficiency complication is the immune dysregulation, followed by organomegaly and recurrent infections. Among patients with immune dysregulation, enteropathy is the most common clinical manifestation, followed by autoimmune hemolytic anemia and idiopathic thrombocytopenic purpura 4. Furthermore, hypogammaglobulinemia is found 57% of the patients. Among patients with hypogammaglobulinemia, 17% of patients had reduced IgM and IgG levels, 17% had reduced IgA and IgG levels, and the 34% had low titers of all three immunoglobulin isotypes. In addition, the 16% patients had reduced IgA levels only, and 16% had low IgG levels 4. To date, no patients with this condition have been reported in Colombia.
The goal of the present study was to evaluate the expression of the LRBA protein in patients with clinical phenotype associated to LRBA-deficiency from Colombia. Subsequently, to evaluate the clinical, immunological characteristics and the possible genetic variants involved in the regulation of the expression of this protein or other associated with an immune system in patients that exhibit decrease LRBA expression.
Materials and Methods
Summary of the methodology is showed in the Figure S1
Human subjects
Patients referred to the Grupo de Inmunodeficiencias Primarias de la Universidad de Antioquia (Medellín, Colombia) presenting with any clinical manifestation of the LRBA deficiency (Immune dysregulation: enteropathy, autoimmune hemolytic anemia, idiopathic thrombocytopenic purpura, granulomatous-lymphocytic interstitial lung disease, type I diabetes and/or chronic autoimmune hepatitis; organomegaly: splenomegaly, lymphadenopathy and/or hepatomegaly; hypogammaglobulinemia: low titer IgM, IgG or IgA levels, or reduced all three immunoglobulin isotypes) were recruited to evaluate the LRBA expression in peripheral blood mononuclear cells (PBMC). Excluded were patients with incomplete diagnosis according to the criteria of the specialist. Healthy controls matched by age and gender with patients, parents and siblings were also included and samples were run in parallel with those of the patients, this was performed to ensure that the variability of expression was not inherent to age or sex and to determine if other members of the family also was affected the expression of the protein. The study was approved by the Institutional Review Board from Sede de Investigación Universitaria (SIU), Universidad de Antioquia, UdeA. Written informed consent was obtained from all participants involved in this study.
Evaluation LRBA expression
Western Blot (WB) Analysis
PBMC were obtained from peripherical blood (PB) isolated by density gradient centrifugation using Ficoll-Hypaque (Sigma-Aldrich, St Louis, MO). PBMC were resuspended in RPMI 1640 medium with L-glutamine and sodium bicarbonate (Sigma-Aldrich), supplemented with 10% of heat-inactivated fetal bovine serum (FBS, Gibco, MA, USA), 2% penicillin/streptomycin mixture (Lonza, Walkersville, MD) (Complete RPMI) and stimulated with phytohemagglutinin A (PHA, 10 mg/mL, Sigma-Aldrich). Cells were harvested and lysed using the RIPA buffer (Cell Signaling, MA) containing a protease inhibitor cocktail (Amresco, GA) and 0.1 M Phenyl methanesulfony fluoride (PMSF, Sigma-Aldrich). Protein levels were measured using the Quick Start(Bradford Protein Assay (Bio-Rad, CA) following manufacturer instructions. Lysates were resolved by SDS-PAGE (Polyacrylamide from Amresco, 10% gradient gel). Proteins were transferred onto polyvinylidenedifluoride (PVDF) membranes (Bio-Rad) and incubated with polyclonal rabbit anti-human LRBA antibody (HPA023597, Sigma-Aldrich), anti-GAPDH (Sigma-Aldrich) and anti-rabbit IgG-peroxidase antibody as secondary antibody (Sigma-Aldrich), were used for the protein detection. Signals were detected by the Western Blotting Luminol Reagent (Santa Cruz Biotechnology, TX) using the Syngene G:Box XR5 chemiluminescent and multicolor fluorescent imaging system (Integrated Scientific Solutions, San Diego, CA). Band quantification was performed using ImageJ 1.48v software (National Institutes of Health, Bethesda, MA). As a control of the reduction of the LRBA expression, the patient's cell lysate CVID022 was used, since it exhibited a decrease in the expression from the first WB performed.
Flow cytometry analysis (Fluorescence-activated cell sorting-FACS)
Ex vivo PB cells and PBMC stimulated w/o PHA (10 µg/mL), were intracellularly stained with the polyclonal anti-LRBA antibody (HPA019366, Sigma-Aldrich) using anti Rabbit PE-F(ab)`2 Donkey IgG (Sigma-Aldrich) as a secondary antibody. Intracellular cell staining was performed using Cytofix/Cytoperm and the Perm/Wash solutions from BD following the manufacturer’s instructions. Cells were re-suspended in Dulbecco PBS 1X (dPBS Sigma-Aldrich) prior to analysis. FACS was performed using a Fortessa II (BD, Franklin Lakes, NJ) and analyzed using the FlowJo V9.9.5 software (Tree Star, Inc. Ashland, OR).
Furthermore, to evaluate LRBA staining specificity from the polyclonal antibody by FACS, the immunization LRBA peptide of the anti-LRBA polyclonal antibody (HPA019366, Sigma-Aldrich) was synthesized by ProteoGenix (Schiltigheim, France) and then diluted in dPBS (Sigma-Aldrich) at a final concentration of 4 mg/mL. Then, different peptide concentrations (2, 0.2, 0.02, 0.002 mg/mL diluted in dPBS) were incubated with the anti-LRBA antibody, and subsequently, specific binding of this anti-LRBA antibody mixture to the cells was evaluated ex vivo intracellular in CD3+ cells from PBMC.
Immunologic studies
Clinical information and immunological analysis (immunoglobulin levels, lymphocyte subpopulation numbers in peripherical blood) from patients were obtained from the Grupo de inmunodeficiencias primarias diagnostic service records.
Whole-Exome Sequencing (WES)
Genomic DNA (gDNA) was isolated from PB obtained from patients and their parents using the DNA Isolation Kit from Puregene™ (Gentra Systems, Minneapolis, MN) following manufacturer’s instructions. gDNA were subjected to WES by Macrogen® (Seoul, Korea), using the Illumina HiSeq X Ten Sequencing System. gDNA were subjected to library preparation, which introduced adapters into the genomic DNA, while fragmenting it into 100 bp segments. The adapter sequence was used to amplify fragmented gDNA in a limited cycle polymerase chain reaction. The DNA was enriched for exonic fragments, which were also amplified5. Sequences were aligned for variant calling and annotation with the human genome reference sequence (hg19 build) using BWA software (http://bio-bwa.sourceforge.net). Downstream processing was performed with Samtools - Faidx and Picard - Dict software, PICARD (http://broadinstitute.github.io/picard/), and the genome analysis toolkit (GATK), https://software.broadinstitute.org/gatk/best-practices/workflow?id=11145.
The command line to select variants in candidate genes was: variants in genes previously associated with PID, population frequency <1% in both 1000 genome and Exome Aggregation Consortium (ExAC), nonsynonymous, located in exonic o splicing regions, deleterious effect predicted by SIFT, Polyphen2, MutationTaster or CADD (Table S1). Homozygote mutations were filtered because we suspected a recessive pattern of inheritance (Fig. S2).
To predict the intronic variants effect, the In-silico tools EX-SKIP was also used (http://ex-skip.img.cas.cz/). Also, NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/) and Fruitfly (http://www.fruitfly.org/seq_tools/promoter.html) predictors were used for testing splice site variants. The reference sequences NM_006726.4 was used for the analysis of the LRBA variants.
Blast Analysis
A BLAST analysis (Basic Local Alignment Search Tool, https://blast.ncbi.nlm.nih.gov) was performed to evaluate if the peptides recognized by the anti-LRBA polyclonal antibodies used in this study (HPA019366 and HPA023597, Sigma-Aldrich) were cross-reacting with other human proteins. The peptides of the anti-LRBA polyclonal antibodies (HPA019366 and HPA023597, Sigma-Aldrich) were subjected to a blast analysis against all reference proteins using the following parameters: Database: refseq protein; Organism: Homo sapiens (taxid:9606); Algorithm: blatstp (protein-protein BLAST).
Statistical analysis
Pearson correlation coefficient using GraphPad Prism 5 (https://www.graphpad.com/support/prism-5-updates/) was performed to measure the statistical correlation between the intensity of the LRBA detection by WB (LRBA band intensity) and the LRBA staining by FACS (Mean fluorescence intensity, MFI).
Results
Detection of LRBA in peripheral blood (PB) cells
To confirm the sensitivity of this test and the accuracy of two different LRBA antibodies commercially available, we standardized the evaluation of LRBA expression in human PBMC by WB and FACS. Using WB, the LRBA band at 319 kDa in PHA-stimulated PBMC cells from healthy donors (HD) was detect (Fig. 1A). Also, LRBA by FACS in ex vivo PB cells and PBMC stimulated w/o PHA. LRBA is constitutively expressed in CD3+ cells from PB ex vivo and in unstimulated PBMC after three days of incubation was detect, however, upon stimulation with PHA, the expression is increased two-fold (Fig. 1B). the correlation between the ability of WB and FACS to detect LRBA was evaluated (Fig. 1C), and a positive correlation (r: 0.4723 p: 0.0112) between the level of LRBA detection among both techniques was observed. To assure the specificity of our antibodies, a BLAST analysis was performed to evaluate if every epitope recognized by the anti-LRBA antibodies was aligning with others human proteins (Fig. 1D). Besides the 100% match with the LRBA protein, these epitopes also matched with the paralog protein NBEA in 36% and 44%, respectively. These results indicate that the polyclonal anti-LRBA antibodies used by us could also recognized NBEA but this possibility is ruled out because this protein has a very limited expression to brain 6. In addition, the antibody Ab HPA019366 showed specific binding to intracellular LRBA that was lost when adding a specific LRBA-blocking peptide in a dose-dependent manner (Fig. 1E).
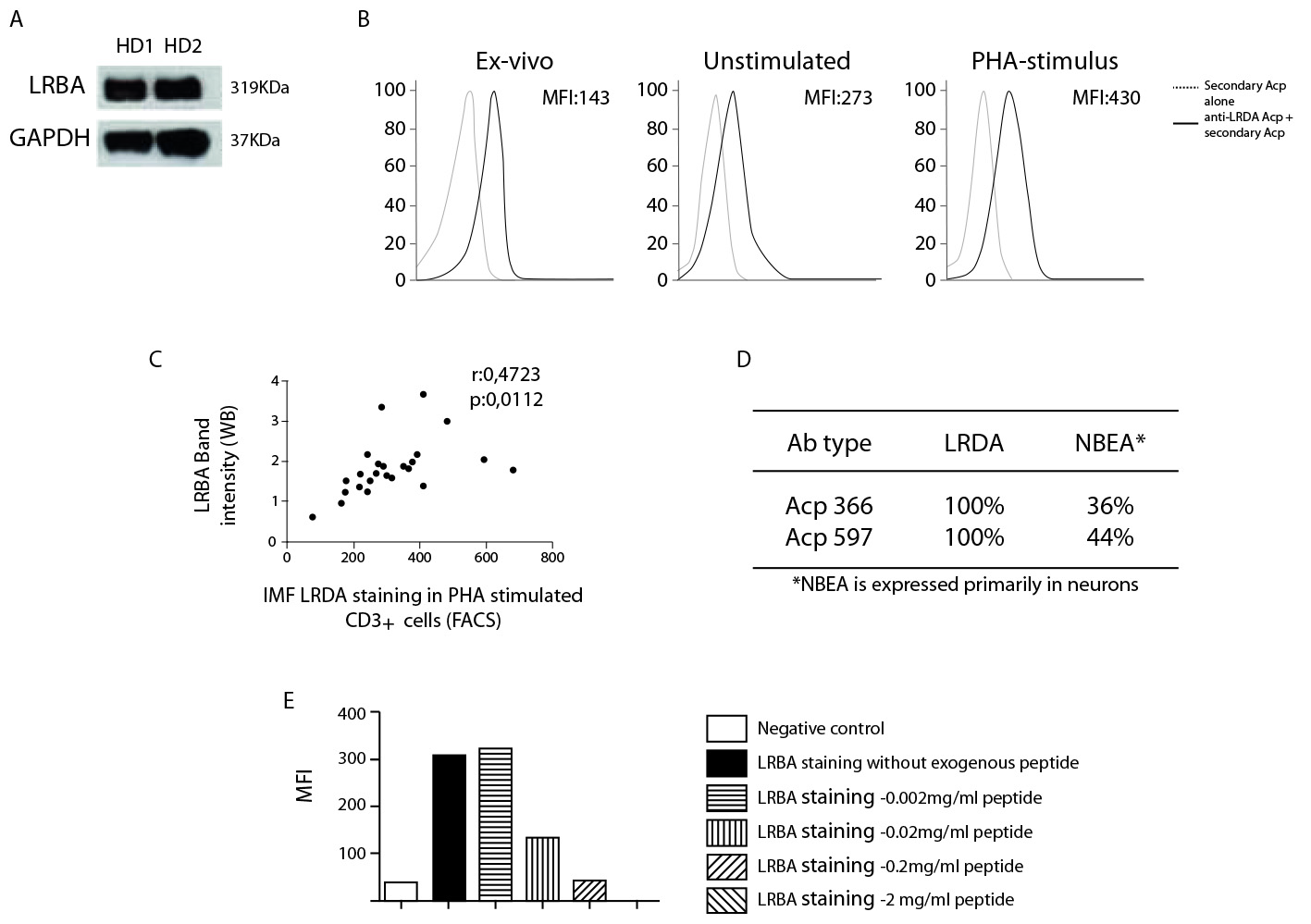
Figure 1 Standardization of the LRBA staining in human peripheral blood cells. Phytohemaglutinin A (PHA)-stimulated peripheral blood mononuclear cells (PBMC) cells from healthy donors (HD) were obtained and immediately lysed to extract and quantified proteins (40 µg) that were tested in a Western blot with the antibody anti-LRBA (HPA023597, Sigma) using GAPDH as a constitutively expressed protein. Bands were detected at 319 and 37 KDa, respectively (A). Consecutively, ex vivo PB cells and PBMC stimulated w/o PHA (1 µg/mL) were intracellularly stained with the antibody anti-LRBA (HPA019366, Sigma) using anti Rabbit PE- F(ab)`2 Donkey IgG as a secondary antibody. Dotted lines indicate the staining only with the secondary antibody. Continued lines represent the staining with the anti-LRBA antibody together with the secondary antibody. Shown is the LRBA staining gating only the CD3+ cells (B). A Pearson correlation analysis was performed by comparing the intensity of the LRBA detection by western blot (LRBA band intensity) and the LRBA staining by FACS (Mean fluorescence intensity, MFI) (C). Also, a BLAST analysis (https://blast.ncbi.nlm.nih.gov) was performed to evaluate if the peptide recognized by the anti-LRBA antibodies used in this study were cross-reacting with NBEA (Sequence from www.ncbi.nlm.nih.gov/nuccore/NC_000013.11) (NBEA is a paralog of LRBA) (D). Considering that anti-LRBA antibodies obtained commercially are policlonal, the specificity of recognition was also evaluated using the LRBA peptide SVLMVSKYRDILEPQNERHSQSCTETGSENENVSLSEITPAAFSTLTTASVEESESTSSARRRDSGIGEETATGLGSHVEVTPHTAPPGVSAGPDAISEVLSTLSLEVNKSPETKNDRGNDLDTKATPSVSV as a competitive reagent at different concentrations. The percentage of the recognition of the cell LRBA protein was calculated (E).
Expression of LRBA in patients with clinical phenotype associated to LRBA-deficiency
According the inclusion criteria, one hundred and twelve patients were recruited, 29 patients with common variable immunodeficiency (CVID), 14 with X-linked agamaglobulinemia (XLA), 55 with idiopathic hypogammaglobulinemia (IHG), 1 with selective IgA deficiency (SIgAD), 5 with enteropathies, 3 with Idiopathic thrombocytopenic purpura and 5 with systemic lupus erythematosus (SLE) (Fig. 2A). In our cohort, patients with CVID, IHG, XLA and SIgAD presented with either hypo or agammaglobulinemia, 9 CVID patients exhibited autoimmune manifestations (Table S2).
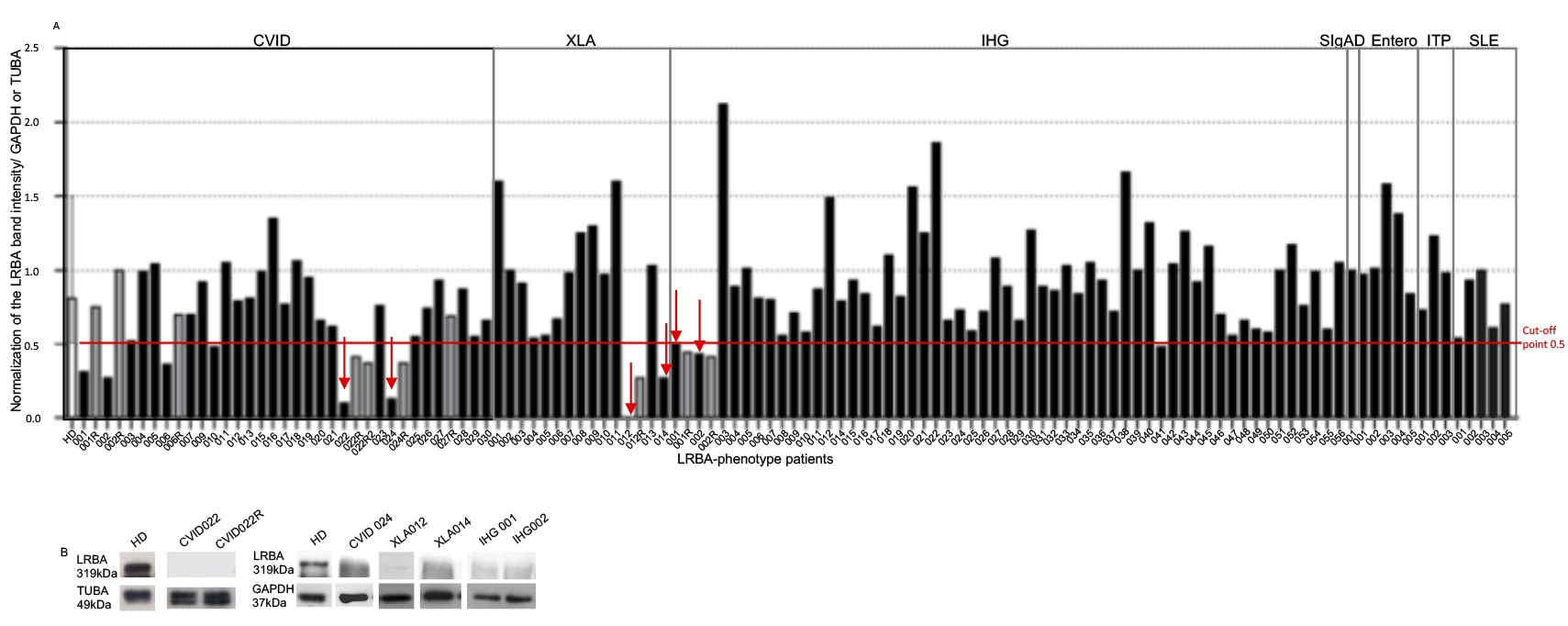
Figure 2 LRBA expression in patients. One hundred and twelve patients were recruited to evaluate the LRBA expression by westerm blot (WB). PHA-stimulated PBMC from 25 healthy donors and patients were obtain and lysed to extract and quantify proteins (40 μg) that were tested in a WB with the anti-LRBA antibody Ab HPA023597 (Sigma) using either GAPDH or TUBA as a constitutively expressed protein. Bands were detected at 319 kDa for LRBA, 37 KDa for GAPDH and 49 KDa for TUBA. Normalization of the LRBA band intensity against the constitutive protein (GAPDH or TUBA) is shown in (A). WB band intensities of LRBA in PHA-stimulated cells from patients with LRBA decreased expression is shown in (B). CVID: Common Variable Immunodeficiency, XLA: X-linked agammaglobulinemia, IHG: Hypogammaglobulinemia, SIgAD: Selective IgA Deficiency, ITP: Idiopathic thrombocytopenic purpura, SLE: Systemic Lupus Erythematosus. The gray bars represent the repeated samples. The red arrows indicate the patients with decrease in LRBA who continued in the study
The relative expression expression of LRBA was evaluated by WB in 25 HD and quantified using densitometry, to normalize it against the expression of the constitutive protein (either TUBA or GAPDH) (Fig. 2A). Optimal cut-of point of LRBA to distinguish between “decrease LRBA expression” and “normal LRBA expression” was selected considering the minimum value of LRBA expression among HD (cut of point 0.5).
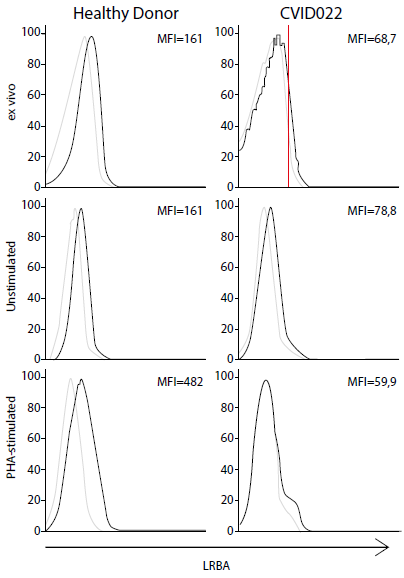
As a negative control, CVID022 was used since this sample was always negative for the expression of LRBA (Fig. 2A). Also, the LRBA expression in ex-vivo PB cells and PBMC stimulated w/o PHA by FACS was evaluated (Figure S3). LRBA exhibited a 2.3-fold decrease in ex-vivo patient´cells compared to that in HD cells. Therefore, the reduction in the expression of LRBA in lymphocytes from CVID022 was observed not only by WB but also by FACS. Unfortunately, it was not possible to evaluate the LRBA expression by flow cytometry in the other patients with decreased LRBA expression by WB. Similar results were observed in PBMC w/o PHA-stimuli. Nevertheless, the effect was more evident in PHA-stimulated PBMC with an 8-fold decrease in the expression of LRBA in the patient as compared to a HD (Figure S3).
Decrease LRBA expression in nine protein lysates from PHA-stimulated PBMC was observed (Figure 2A, gray bars), but only were repeated in eight and confirmed in six lysates by obtaining a second blood sample: two CVID patients (CVID22 and CVID024), two XLA patients, (XLA012 and XLA014) and two hypogammaglobulinemia patients (IHG001 and IHG002) (Figure 2 red arrow). Interestingly, the hypogammaglobulinemia patients are siblings. It is important to mention that CVID027 is a sister of these siblings, however she did not show decrease LRBA expression in PBMC (Figure 2A).
There was no difference in the expression of LRBA by WB in cells obtained from patients with the different clinical conditions evaluated in comparison to HD (Mean/SD of the normalized band intensity: HD: 0.86/0.25; CVID: 0.69/0.32; XLA: 0.86/0.47; IHG: 0.90/0.35; enteropathies: 1.1/0.31; ITP: 0.98/0.25 and SLE: 0.77/0.2).
Interestingly, an increase in the LRBA expression in 7 patients was observed (XLA001, XLA011, IHG003, IHG022, IHG022, IHG038, Entero003). The Figure 2B show WB band intensities of LRBA in PHA-stimulated cells from patients with LRBA decreased expression
Clinical phenotype of patients with decreased LRBA expression
The clinical characteristics of the patients is showed in the Table 1. The CVID022 patient is a female born full-term in 1982, product of the first pregnancy to unrelated healthy parents from Medellin (Antioquia-Colombia). Although no family history of recurrent infections, autoimmunity or hematological malignancies was documented, rhinitis, bronchitis, diabetes type II, asthma and chronic obstructive pulmonary disease were described in the family members.
Table 1 Summary of the clinical characteristics of patients with decreased LRBA expression
| Patient | Current Age (y) | Age at first symptom (y) | Age at diagnosis (y) | Recurrent Infections | Gastrointestinal diseases | Other clinical manifestations | |
|---|---|---|---|---|---|---|---|
| Allergies | Anemia | ||||||
| CVID022 | 36 | 10 | 13 | Tonsillitis, pneumonia, otitis, sinusitis. | Diarrhea with mucus (giardiasis and amebiases) gastritis and duodenal polyps | Rhinitis | Yes |
| CVID024 | 19 | 2 | 35 | Tonsillitis, pneumonia, otitis, parotitis, periorbital cellulitis, sinusitis. | Appendicitis with abdominal sepsis, irritable bowel disease | Conjunctivitis, Rhinitis | - |
| XLA012 | 7 | 1 | 1 | Urinary tract infection, otitis, Aspergillosis†, chronic bronchial syndrome, sinusitis. | Acute gastroenteritis and diarrheal disease (Salmonellosis) | - | Yes |
| XLA014 | 4 | 1 | 1 | Upper respiratory tract infections, otitis, sinusitis. | - | Rhinitis | - |
| Hipo001 | 9 | 1 | 3 | Bronchiolitis, pneumonia, otitis media, laryngitis | Gastroesophageal reflux disease (GERD) grade IV, food allergy | Asthma | - |
| Hipo002 | 11 | 1 | 4 | Otitis, tonsillitis | Gastrointestinal infection, diarrhea, chronic abdominal pain | - | - |
† This infection was systemic but not recurrent.
The CVID024. patient is a male, born full-term in 1998 to unrelated healthy parents. He is the first child and has a younger healthy brother. His father is from Medellin and his mother from Bolivar (Antioquia-Colombia). He only had a family history of type II diabetes and asthma in his paternal grandmother. At age 8, two variants in TACI were identified in a compound heterozygous pattern of inheritance: One variant, c.121G>C (p.41D>H) in exon 2 and the second variant c.297_299insT (p.Q99fsX6) in exon 3. However, both variants were also identified in the unaffected mother and brother of the patient. The father was found wild type for both variants.
The XLA012.patient is a male, born by C-section in 2010 to unrelated healthy parents from Cúcuta (Norte de Santander, Colombia). He has also a healthy older sister. Diabetes and hypertension were reported in his maternal grandmother. Familiar history of a deceased uncle at the age of 5 due to “edema” was also documented. The genetic diagnosis of XLA was performed at age 2 with the detection of the mutation c.1573C>T p.Arg525X in Btk. During the five years follow up, a chronic bronchial syndrome, anemia and chronic sinusitis has been documented.
The patient XLA014.is a male, born full-term in 2013, product of the second pregnancy to unrelated healthy parents from Santa Rosa del Sur de Bolivar (Santander-Colombia). His mother had a previous spontaneous abortion and he has also a healthy younger sibling. Hematological cancer was reported in his paternal grandmother.
IHG001 is a male, born in 2008 in Medellin - Colombia from unrelated healthy parents. He was the product of the third pregnancy, born prematurely at 34 weeks of gestation by C-section with hyaline membrane disease. His father suffered from allergic rhinitis and his maternal grandmother documented asthma and hypothyroidism. He has also a cousin with hypothyroidism. His sister (IHG002) was born in 2006. She was the product of the second pregnancy, born at 36 weeks of gestation by C-section. These siblings have the older sister (CVID027) who had normal serum immunoglobulins and LRBA expression in PBMC.
Immunological phenotype in patients with decrease LRBA expression
An immunological analysis of the different lymphocyte subsets in PB from all patients with LRBA decrease in PBMC did not show a common and distinctive phenotypic feature (Table 2). However, the immunological characteristics from the different patients were confirmed according to the disease group. All patients with CVID exhibited a serum IgG and IgA decrease. The T cell subpopulations in PB were normal in the CVID patients. A decrease in the memory B cells (IgD+/CD27+) was observed in both patients however, only CVID022 had a decreased in the switched memory B cells (IgD-/CD27+). With regard to the XLA patients, the decrease in the three serum Ig isotypes was observed in XLA012 whereas no serum Ig data previous to the IVIG replacement therapy was found for XLA14. Notorious was the very low numbers of PB B cells observed in these patients. Also, some minor alterations of CD3+ T cells were observed in the XLA patients. Interestingly, the two siblings with IHG exhibited different immunological phenotype. IHG001 exhibited very low levels of serum IgG and IgA whereas IHG002 presented with only decreased serum IgG. However, both exhibited increase in the memory B cell subpopulation (Table 2).
Table 2 Immunological characterization of the patients with decreased LRBA expression in PHA-stimulated PBMC
| Patients | ||||||
|---|---|---|---|---|---|---|
| CVID022 | CVID024 | XLA012 | XLA014 | IHG001 | IHG002 | |
| Immunoglobulin levels (mg/dL) | ||||||
| IgG | 0 ↓ | 19.2 ↓ | 111.8 ↓ | ND | 6.4 ↓ | 675.4 ↓ |
| IgM | 5.7 ↓ | 55.1 ↑ | 53.1 | ND | 64.0 | 82.0 |
| IgA | 2.1 ↓ | 2 ↓ | 5.2 ↓ | ND | <3.3 ↓ | 72.2 |
| Lymphocytes | Percentages/Absolute numbers (cells/mm3) | |||||
| CD3+ | 64.0/1,169 | 74.0/2,963 | 90.5/2222 ↑ | 88.0/4,738 ↑ | 64.4/2,655 | 74.0/2,776 |
| CD3+ CD4+ | 44.8/524 | 23.4/937 | 56.7/1,393 ↑ | 25.6/1,366 | 35.2/1,452 | 35.8/1,346 |
| CD3+ CD8+ | 52.0/606 | 40.2/1,610 | 27.4/673 | 58.3/3,111 ↑ | 23.3/961 | 31.8/1,196 |
| CD19+ | 8.2/147 ↓ | 12.8/230 | 0.7/16 ↓ | 0.1/3 ↓ | 23.3/961 ↑ | 17.6/660 |
| CD19+/CD21+ | 7.0/125 | 11.0/197 | 0.4/11 | 0.0/1 | 22.1/914 | 16.1/606 |
| CD19+CD27+ | 14.8/22 ↓ | 16.0/37 | -- | -- | 24.8/238 ↑ | 30.6/202 ↑ |
| IgD+/CD27- | 77.2/113 ↑ | 68.0/156 | -- | -- | 71.7/689 ↑ | 63.6/420 ↓ |
| IgD+/CD27+ | 10.5/15 ↓ | 5.9/14 ↓ | -- | -- | 15.1/145 ↑ | 15.6/103 ↑ |
| IgD-/CD27+ | 4.3/6 ↓ | 10.1/23 | -- | -- | 9.7/93 ↑ | 15.0/99 ↑ |
| IgD+/CD38+ | 0.5/1 | 12.9/30 | -- | -- | 11.1/107 | -- |
| IgD-/CD38++ | 0.0/0 ↓ | 3.3/2.1 | -- | -- | 0.93/9 ↑ | 1.5/10 |
| CD24++/CD38++ | 0.5/1 ↓ | 12.8/29 | -- | -- | 11/106 | 13.0/86 |
Ig: Immunoglobulin, ND: no data. The arrows down indicate decrease and the arrows upwards with respect to the reference value.
LRBA variants in the patients with decrease LRBA expression.
WES was performed in the patients with decrease LRBA expression. Overall, eighteen either non-synonymous exonic or intronic LRBA variants were identified in these patients (Table 3). Five variants with population frequency (1% were found (Table 3, gray bars), which two possibly pathogenic variants rs148385798 in CVID022 patient and the rs950337550 in CVID024 patient were found (Table 3, red box). The variant rs148385798 is a heterozygous missense change c.T194C in the exon 2 which correspond to the change p.I65T in the ConA domain of the protein. The In silico analysis predicted to be “deleterious” with SIFT (score: 0.02); “disease causing” with MutationTaster (p-value: 0.995) and “deleterious” in PROVEAN (score: -2.558) and CADD score was 16.37. The second variant rs950337550 is the heterozygous missense variant c.6892A>G in exon 46 which confers a p.K2298E change in the BEACH domain. The in silico analysis predicted to be “deleterious” with SIFT (score: 0.005); “disease causing” with MutationTaster (p-value: 1) and “Non-deleterious” with PROVEAN (score: -1.44).
Table 3 LRBA variants in the patients with decrease LRBA expression in PHA-stimulated PBMC
| Patients | Location | Ref | Alt | Exons/introns | SNP | Alleles | Frequency |
|---|---|---|---|---|---|---|---|
| CVID022 | 151199080 | G | A | Exon | rs2290846 | het | 0.15 |
| 151207127 | C | T | Exon | rs3749574 | het | 0.16 | |
| 151242312 | C | T | Intron | rs11737450 | het | 0.18 | |
| 151753134 | A | - | Intron | rs145452262 | het | 0.062 | |
| 151765243 | G | A | Intron | rs186080 | hom | 0.75 | |
| 151935601 | A | G | Exon | rs148385798 | het | 0.002 | |
| 151408800 | T | C | Intron | rs7674989 | hom | 0.2 | |
| 151656589 | C | T | Intron | rs1993109 | het | 0.4 | |
| 151829659 | T | C | Intron | rs1201207 | hom | 0.04 | |
| 151773593 | G | C | Exon | rs1782360 | hom | 0,29 | |
| XLA12 | 151207127 | C | T | Exon | rs3749574 | het | 0.16 |
| 151236393 | A | G | Intron | rs74459799 | het | 0.0062 | |
| 151242312 | C | T | Intron | rs11737450 | het | 0.18 | |
| 151357938 | G | A | Exon | rs950337550 | het | 0.000008 | |
| 151765243 | G | A | Intron | rs186080 | hom | 0.75 | |
| 151817507 | C | T | Intron | rs1813134 | het | 0.011 | |
| XLA14 | 151765243 | G | A | Intron | rs115139393 | hom | 0.75 |
| 151829659 | TC | - | Intron | 151829659 | hom | 0.96 | |
| 151231371 | T | C | Intron | rs1813134 | het | 0.52 | |
| 151187011 | - | GAGAT | Intron | rs201486611 | het | 0.69 | |
| IHG001 IHG002 | 151231371 | T | C | Intron | rs1813134 | hom | 0.52 |
| 151235987 | G | A | Intron | -- | het | -- | |
| 151753134 | A | - | Intron | rs145452262 | het | 0.062 | |
| 151762104 | G | A | Intron | rs893110507 | het | 0.00004 | |
| 151765243 | G | A | Intron | rs186080 | het | 0.75 |
Ref: allele reference, Alt: allele altered, SNP: single nucleotide polymorphism, hom: homozygous, het: heterozygous. Highlighted in gray are the variants with a frequency <0.001 in the 1,000 genomes project (http://www.internationalgenome.org). The variables indicated in red are pathogenic or possibly pathogenic according to the predictors in silico.
Considering that LRBA inheritance pattern is autosomal recessive and that we had not found biallelic variants, we continued to the variants analysis in other genes involved in the immune system.
Other relevant variants found by Whole Exome Sequencing (WES) analysis
Were identified 50,300 high-quality variants in the CVID022 exome. Among them, 17056 were homozygous and 33244 heterozygous (Table 4). The variants in PID genes were first analyzed and homozygous variants in three genes (PLCG2, POLE, RLTPR) were found. However, these variants were localized in intronic regions, very distant from the proximal exon. To proceed with the WES analysis, following the hypothesis of a recessive mood of inheritance, homozygous or compound heterozygous variants were selected. These variants should fulfill also the defined criteria. Three candidate genes (KCNN3, TRPM3, ECT2L) were selected, however KCNN3 and TRPM3 are not expressed in lymphoid tissues. Moreover, the patient’s father also carried both variants in ECT2L. Finally, we evaluated the de novo heterozygous variants. One variant in Mitochondrial Ribosomal Protein L3 (MRL3) was found. Diseases associated with MRPL3 include Combined Oxidative Phosphorylation Deficiency 9 7. however, this is a severe condition that primarily impairs neurological and liver function.
Table 4 Summary of whole-exome sequencing results in patients with decrease LRBA expression
| Variants | CVID022 | XLA012 | XLA014 | IHG001 | IHG002 |
|---|---|---|---|---|---|
| Total number | 50,300 | 53,002 | 52,821 | 53,215 | 53,215 |
| Number of Homozygous | 17,056 | 14,785 | 15,015 | 14,654 | 14,654 |
| Number of Heterozygous | 33,244 | 20,731 | 20,530 | 21,352 | 21,352 |
| PID related | PLCG2, POLE, RLTPR | BTK | BTK | --- | |
| Homozygous candidates | KCNN3, TRPM3 | --- | --- | PLCL2, FDFT1, GPRIN2 | |
| Heterozygous candidates | ECT2L | --- | --- | SEC22B | |
In XLA12, 53,002 variants were identified, 14,785 homozygous and 20,730 heterozygous (Table 4). Within the PID genes, 27 variants were identified. Among them, the variant c.1573C>T p.Arg525X in BTK was confirmed. In other genes, no potential pathogenic candidate variants were found.
In XLA14, 52,821 high-quality variants were identified in the exome, 15,015 were homozygous and 20,530 heterozygous (Table 4). Within the PID-related genes, 40 variants were found and one variant potentially pathogenic one in BTK gene was identified. This correspond to c.622+1G>A (NM_001287344). 3. An in silico analysis (fruit fly and NetGene2) showed that the effects of the BTK mutation conferring a change in the splice donor site at the end of the intron 6 within this gene affecting the protein at the PH domain.
In the IHG001 and IHG002 siblings, a total of 53,215 variants were found in both. Homozygous and heterozygous variant numbers are specified in Table 4. However, no common homozygous or compound heterozygotes in PDI genes were identified. Homozygous variants in PLCL2, FDFT1 and GPRIN2, however, their father had the same homozygous variants in PLCL2 and FDFT1 and their mother had same homozygous variants in GPRIN2. Also, compound heterozygotes variants in SEC22B were found, however, their father carried the same variants.
Discussion
In this work, the expression of LRBA in 112 patients with clinical manifestations associated with LRBA-deficiency was evaluated. The inter-individual and intra-individual concordance in detection of LRBA expression in PHA-stimulated PBMC was observed. LRBA expression as detected by WB, was very heterogeneous in HDs. This heterogeneity has also been reported by Gamez et al. (8, who found by FACS, a MFI range between 2.14 - 10.00 (min-max) in the relative expression of LRBA in PBMC upon stimulation with PHA for four days in HD. Among these patients, decrease LRBA expression in nine protein lysates from patients was observed, after repeated, only in six protein lysates from patients was observed decrease. We still do not know why cells from some patients exhibited discordant LRBA expression levels in different blood samples and time points. A possible explanation for these results is an immune-triggered cellular senescence. Different factors such as persistent viral infections, stress and inflammatory syndromes induce the accumulation of T cells with characteristics of senescence 9. It is possible that these patients have been going through periods of stress or infection with the consequence of an induced senescence and alterations in the proliferation of the cells. Since the detection of LRBA expression in PBMC in our settings is an activation-dependent phenomenon, decrease activation of senescent cells could lead to a decrease in the immune cell-detected expression of LRBA. However, an activation marker, such as CD69, should be included in the staining controls in the future studies. On the other hand, an increase in the LRBA expression in 7 patients was also observed. To date, overexpressed of this gene associated with some PID has not been reported, however, Wang et al. (10, in 2004 reported by microarray and real time PCR analyses that LRBA is overexpressed in several cancers relative to their normal tissue controls. Additionally, the expression of cell activation must have been monitor in our patient’cells, since it has been previously shown that LRBA expression increases upon cell activation 4. This could be stated as one of the limitations of our results since decreased LRBA expression by WB might have been caused by poor immune cell activation. However, we had always sample limitation due to the enormous amount of protein required to perform the WB. Moreover, there has been no correlation among the intensity of the LRBA expression by flow and the proliferation index in B cells in healthy donors (Data not shown). On the other hand, patients with poor proliferation of T and B cells included in this cohort, exhibited normal LRBA expression by WB (Data not shown).
The patients with decrease LRBA expression exhibited different phenotypic and immunologic characteristic. However, all patients presented with a reduction in IgG and recurrent infections and all except one presented gastropathy. Pathogenic or potentially pathogenic biallelic variants neither in LRBA nor in other immune-related genes were not observed. LRBA expression in PBMC may be decreased due to epigenetic mechanisms 11. Epigenetic mechanisms determine the phenotype without changes in the genotype, which operate at the transcriptional and post-transcriptional level of gene activity as well as at the level of protein translation and post-translational modifications. They participate in such processes as cell differentiation, morphogenesis, variability and adaptability of an organism and may be affected by both genetic and environmental factors 11. Moreover, epigenetic changes have been proposed to contribute to the CVID pathogenesis. Rodríguez-Cortez et al. (12, demonstrated by high-throughput DNA methylation analysis, in monozygotic twins discordant for CVID, predominant gain of DNA methylation in B cells in the CVID patients with respect to those from the healthy siblings. Taking together, the application of epigenetic studies to identify and characterize not only molecular signatures of LRBA deficiency as well as the mechanisms underlying disease pathogenesis and responses to new therapeutic modalities are required. Another possible explanation of the decrease in the expression of LRBA in PHA-stimulated PBMC from patients with no mutations in either LRBA or other genes by WES, would be the presence of pathogenic mutation in regulatory elements, including promoters, enhancers, insulators, and silencers in LRBA. Any variant that is located within a functional genomic region has the potential ability to cause a dysregulation on gene expression through modifying regulatory elements, possibly resulting in diseases pathogenesis 13. In recent years, genome-wide association studies (GWAS) identified over ten thousand variants associated with various diseases/traits, ~90% of which localize outside of known protein-coding regions 14. In addition, to evaluate the copy number variation and coverage rate of every LRBA exon, to exclude a second heterozygous variant in patients with high clinical suspicion of LRBA deficiency, are required in future studies.
In the present study, variables with clinical significance in LRBA were not found, then, we continued with the analysis of variables in other genes throughout the exome. A new variant (c.622+1G>A NM_001287344) in BTK that probably confers a lost in the splice donor site at the end of the intron 6 in the XLA14 patient was found. When a naturally-occurring splice-sites are abolished by mutations, the spliceosome usually either employs the next available legitimate splice-site (exon skipping) or selects the next best, albeit illegitimate, splice-site in the vicinity (cryptic splice-site utilization) 15. In this specific case, the mRNA transcription might be impaired after exon 6, affecting the protein from the amino acid 25 and generating a new truncated protein. The aberrant mRNAs could be removed by nonsense-mediated decay pathway, which targets mRNAs harboring premature termination codons for degradation 16. Thus, the truncated protein, might not contain the PH, SH3, SH2, and protein kinase domains of the protein and therefore, might be either not functional or subjected to degradation by the Ubiquitin-Proteasome Pathway. However, the exact effect of the mutation should be verified in functional studies.
Taking together, the results from this study indicate, LRBA expression varies greatly between different HDs and patients PBMC. Despite the great variability in the LRBA expression in the different samples, six patients exhibited a decrease expression of this protein by WB in PHA-stimulated PBMC. No genes were detected associated with the defective LRBA production by these cells in these patients. Homozygous mutations in LRBA cause LRBA-deficiency affecting the protein expression. However, reduction LRBA-expression in patients without homozygous mutations or in associated genes were present. These results suggest the other genetic, epigenetic or environmental mechanisms, that might be regulated the LRBA-expression.
The specificity of the LRBA detection by WB and FACS was previously evaluated. A positive correlation between the ability of WB and FACS to detect LRBA was demonstrated. FACS experiments using competitive epitopes to validate the specificity of recognition of LRBA were also performed. With these results, we demonstrated that antibodies showed specific binding to intracellular LRBA that was lost when adding a specific LRBA-blocking peptide in a dose-dependent manner.











 text in
text in