Remark
| 1) Why was this study conducted? |
| To evaluate a possible hereditary genetic factor related to the development of Glaucoma in a family group. |
| 2) What were the most relevant results of the study? |
| Four pathogenic molecular variants in genes associated with glaucoma were identified using massive exome sequencing techniques, which were: rs11938093 in the BTC gene; rs7336216 in the CENPJ gene; rs3817672 in the TFRC gene and, rs983034 in the WLS gene. |
| 3) What do these results contribute? |
| The study provides new information on possible pathogenic molecular variants in genes associated with glaucoma pathology, which will serve as a reference for future molecular epidemiology studies that calculate the risk of developing the disease by carrying these variants. |
Introduction
Glaucoma is the leading cause of irreversible blindness worldwide. This disease consists of the development of a chronic progressive optic neuropathy characterized by increased cupping of the optic disc, associated with progressive vision loss. This disease is related to multiple risk factors such as increased intraocular pressure; diagnosis of arterial hypertension and diabetes, ethnicity; and age 1. It is estimated that there are currently more than 67 million patients affected by glaucoma, estimating a worldwide prevalence of over 15% for Afro-descendant communities, of which 10% present bilateral blindness 2,3. One of the most complex aspects of this disease is the gradual development of its symptoms. Vision loss is progressive and irreversible, and visual field loss is usually only recognized by the patient in advanced stages. Although the determination of intraocular pressure is the most appropriate screening method in cost-effective terms for the identification of suspected cases of glaucoma, it is not enough to establish the diagnosis of the disease. There must be evidenced at least one other clinical criterion that demonstrates damage of the optic nerve, such as increased cupping of the optic nerve, visual field loss, or the detection of hemorrhages at the level of the optic disc 4.
In developed countries, it is estimated that more than 50% of people with glaucoma have not been diagnosed; in developing countries, this lack of diagnosis could be even higher, in the order of 60 to 80% 5,6. It is suggested that up to 90% of cases of blindness due to glaucoma could be avoided through early detection and timely pharmacological treatment 7.
In previous studies in six Colombian cities where the prevalence of glaucoma was estimated in patients with hypertension and diabetes 8,9, a Raizal family with a history of visual diseases was identified in the archipelago of San Andrés and Providencia, where some of their members were diagnosed with glaucoma. This suggests a possible hereditary genetic factor related to the development of this type of pathology in this family group.
In the present investigation, a whole exome analysis was carried out to identify potentially harmful non-synonymous single nucleotide molecular variants in genes associated with glaucoma pathology in members of a Raizal family from San Andrés and Providencia with a history of presenting visual diseases.
Materials and Methods
Type of study
A cross-sectional descriptive study of exome analysis was carried out in a family group with a history of glaucoma. The previous diagnosis of glaucoma was made at the Lynd Newball ophthalmology clinic in San Andres Islas, Colombia. The research had the approval of the institutional review board of the Health Faculty of Universidad del Valle, endorsement numbers: 106-019 of the year 2019.
Study participants
The participants were seven members of a Raizal family with a history of glaucoma pathology who reside in the archipelago of San Andrés and Providencia. Four of the family members had a positive diagnosis for glaucoma, one had a suspected diagnosis, and two were negative. There were also invited to participate two Raizal people who did not belong to the family group and had a negative diagnosisof glaucoma. All participants were of legal age and agreed to participate by reading and signing informed consent. The names of the participants were anonymized and replaced by a numerical code.
Sampling
A professional from the health area performed the venipuncture in the cubital fossa with a 21-gauge needle and a 4ml BD Vacutainer® tube collection system, anticoagulated with 7.2 mg of K2 EDTA. The blood samples were stored at -20° C and transported to the molecular pathology laboratory of Universidad del Valle.
DNA extraction and whole exome sequencing
The QIAamp DNA blood mini kit from QIAGEN was used for DNA extraction, following the manufacturer's protocol. DNA concentration and purity were evaluated in a Nanodrop® ND-2000c spectrophotometer (Thermo Scientific®). The optimal parameters for sequencing were: a DNA concentration greater than 10 ng/uL, and an absorbance ratio ≥1.8 (260 nm/280 nm) and ≥2 (260 nm/230 nm). Additionally, DNA integrity was evaluated by 0.8% agarose gel electrophoresis.
SeqCap EZ Exome kits from Nimblegen V2.0 and TruSeq from Illumina were used for exome enrichment. Libraries were constructed with ~300 bp insert lengths and sequenced on an Illumina HiSeq-2000 (Illumina, San Diego, California). The readings obtained were 150 bp in Paired-End mode with 100X coverage.
Bioinformatic analysis
For quality control, raw data was analyzed using FastQC v0.11.7 software. The sequence filter was performed with the BBMap_38.25 10 program and the FASTX-Toolkit v. 0.0.13 (http://hannonlab.cshl.edu/fastx_toolkit/). The reads were mapped using the human genome GRCh38.p12 as a reference with the BWAv.0.7.17 software and the BWA-MEM algorithm 11. The alignment files were processed using SAMtools 12. As part of post-alignment processing, PCR duplicates were marked using Picard's MarkDuplicates.jar tool and removed from further analysis (https://github.com/broadinstitute/picard). Using the GATK HaplotypeCaller, version 4.0.2.1 13, it was performed molecular variant calling of single nucleotide variant and InDels simultaneously in Genomic Variant Call Format. For the Genomic Variant Call Format filter, it was used VCFtools v.0.1.16. The single nucleotide variants and InDels were annotated using ANNOVAR v.2018 14, incorporating information from NCBI (Homo sapiens Annotation Release 109) and UCSC genome browser 15.
Genetic ancestry analysis
Genetic ancestry components were estimated for each participant from a panel of 250 ancestry informative markers following the protocol proposed by Wang et al. 16. Genetic ancestry was inferred with the programs: vcftools 17; bcftools 18; tabyx 19; and structure 20, available in the R package. The ancestry informative markers of each participant were filtered with those of 1,305 individuals belonging to three different continental reference populations of the 1,000 genomes project: Africa, Europe, East Asia / African, European, and East Asian. For the grouping of the participants, a principal component analysis was carried out using the "read.vcfR" function of the R package 20. From the missMDA program with the Admixture_gt2PCA format command, the haplotypes of each loci were converted to a numerical code and the missing data were imputed with the estim_ncpPCA command 21. With the dudi.pca command from the ade4 package, the principal components analysis 22 was obtained. Finally, the principal components analysis results were confirmed by means of a Bayesian analysis to infer the ancestral proportions of each individual by means of the Structure program 20. The results were represented by the percentage of ancestry of each population of origin for each individual using bar graphs with the "ggplot2" package 23.
Prediction of harmful non-synonymous single nucleotide variants
Non-synonymous single nucleotide variants molecular variants were filtered from the Genomic Variant Call Format file, taking into account only those with a reference code (rs). Using the JVenn program (http://jvenn.toulouse.inra.fr/app/example.html), non-synonymous single nucleotide variants present in the family group were identified and compared with the two non-related Raizal participants. Subsequently, the variants were filtered according to the gene they affected, using the list of molecular variants associated with the development of glaucoma, which is available in the free access repository DISGENET (https://www.disgenet.org/Glaucoma /CUI_C0017601). Damage prediction of non-synonymous single nucleotide variant variants in protein structure and function was established using the Combined Annotation Dependent Depletion_CADD algorithm 24,25. The CADD value for each single nucleotide variant was obtained from the free access server SNP-NEXUS (https://www.snp-nexus.org/). Subsequently, this value was compared with the score calculated by the Mutation Significance cutoff server to establish the prediction of damage to the structure or function of the protein (https://lab.rockefeller.edu/casanova/MSC). Great damage was considered when the CADD value in the single nucleotide variants was more significant than that calculated for the gene with the Mutation Significance cutoff; otherwise, it was considered as low damage 26.
Results
Figure 1 represents the family relationships among the members of the Raizal family and the study participants who underwent exome analysis. Four of the seven participants belonged to the third generation and three to the fourth, according to the heredogram constructed from the information collected by interviewing family members. Of the four participating individuals of generation III, two groups of siblings were identified: participating individuals III-5 and III-7, half-siblings of the same father but different mother; and participating individuals III-14 and III-15. Except for III-4, all were diagnosed with glaucoma. The parents of these two groups were identified as siblings; and both had been diagnosed with glaucoma: individuals II-2 and II-4.
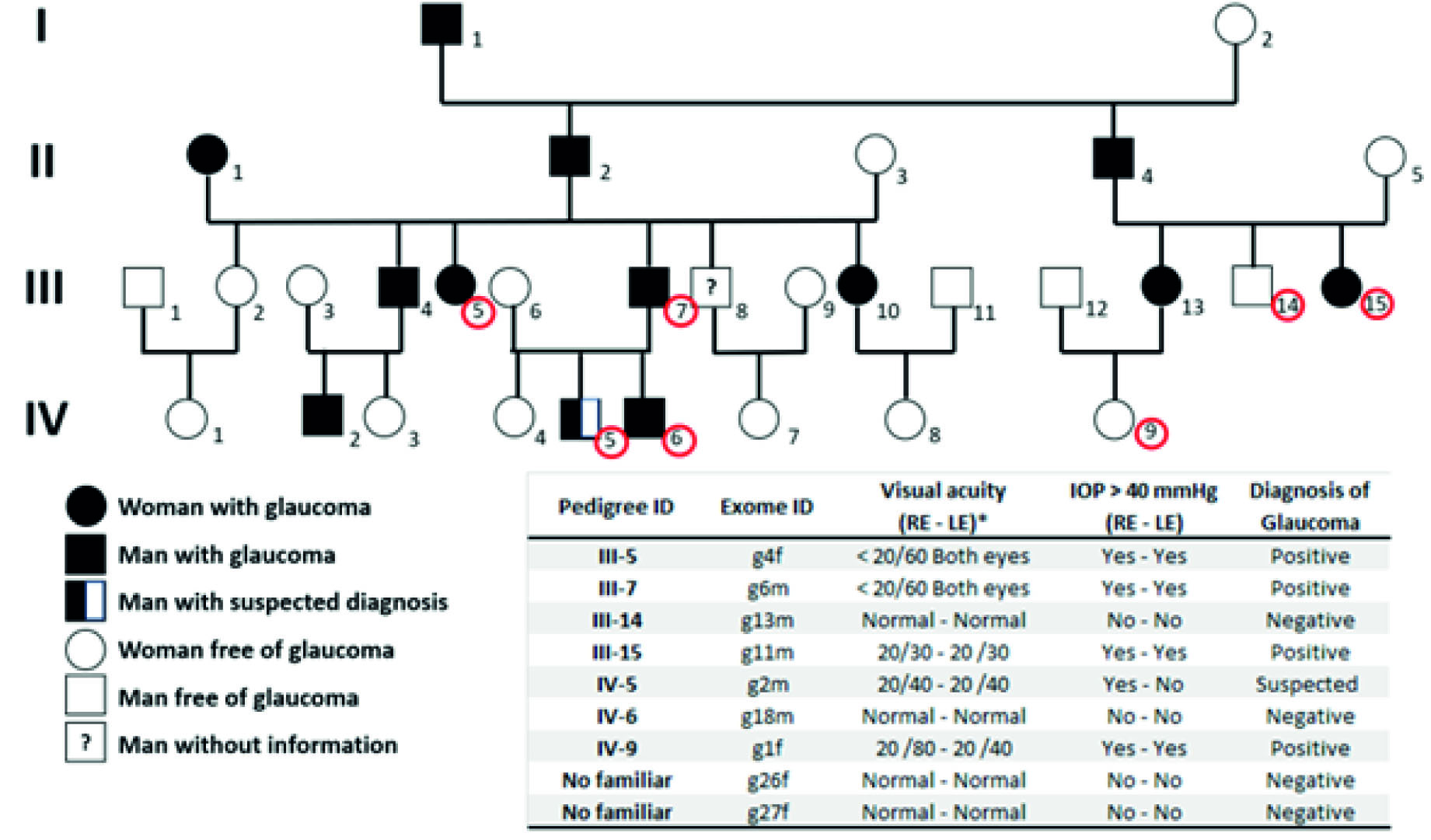
Figure 1 Heredogram of a Raizal family members from San Andres, Providencia, and Santa Catalina with a history of glaucoma. (*) RE= right eye; LE= left eye. Normal visual acuity is equal to 20/20. IOP = intraocular pressure. Red circles identify study participants. Two non-family-members Raizal participants (g26F and g27F) with no diagnosed visual diseases were included in the analysis to identify possibly pathogenic molecular variants. The last column shows the glaucoma diagnoses (as positive or negative ones).
Regarding the IV generation, the participating individuals IV-5 and IV-6 were brothers, sons of III-7. Of this group of siblings, IV-5 was suspected of (presenting / suffering glaucoma. In this same generation participated individual IV-9, diagnosed with glaucoma, whose mother, individual IV-13, had also been diagnosed with glaucoma. IV-5 was the niece of III-14 and III-15 (Figure 1).
Figure 2 shows the percentage estimate of the ancestral genetic components of the study participants. In the principal component’s analysis, it was found the separation of the three ancestral populations: the separation of the African from the European and East Asian with a PC1 equal to 37.8%, and the separation of the European from the African and East Asian with a PC2 equal to 24.8%.
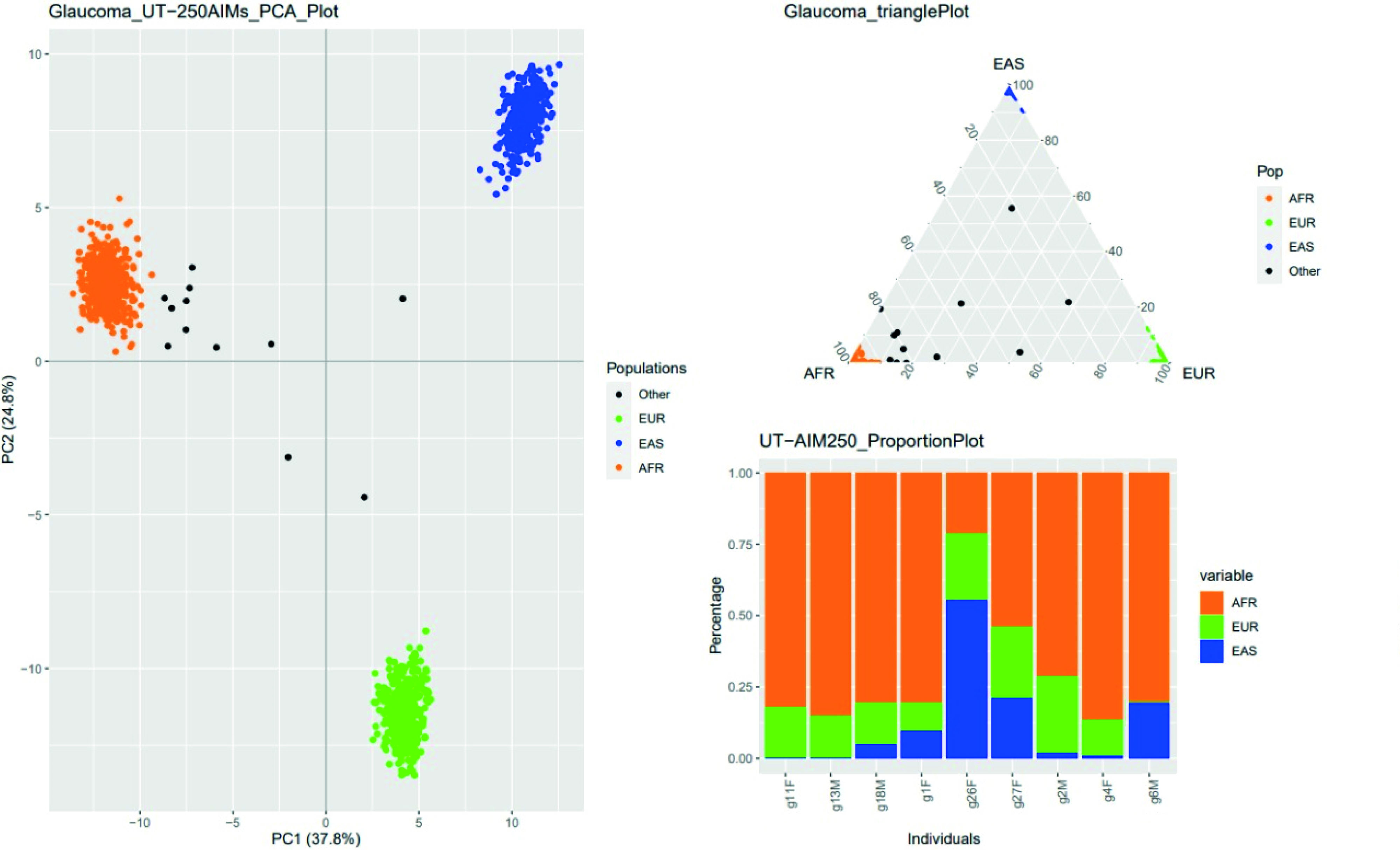
Figure 2 Ancestral genetic components of Raizal individuals from San Andres, Providencia and Santa Catalina. The estimate was made from the analysis of 250 informative markers of ancestry. Each participating individual was compared with data from 1,305 subjects from three distinct continental populations: African (AFR), European (EUR), and East Asian (EAS), available in the 1,000 Genomes project database. N.F.= Not familiar.
Six of nine Raizal people presented a mixture of ancestral genetic components of African, European and East Asian origin; two with African and European origin; and one with African and East Asian genetic ancestry. However, the predominant ancestral genetic component was African, with a percentage greater than 50%, except for one participant who did not belong to the Raizal family, N.F_g26F, who presented an ancestral component mainly from East Asia, with a percentage greater than 50% (Figure 2).
Table 1 shows the number of non-synonymous single nucleotide variants and InDels identified in the exomes of study participants. The average number of non-synonymous single nucleotide variants identified in the members of the Raizal family was 12,366 ±190; and in the two Raizal participants not related to the family, it was 11,772 ±512. In relation to the InDels, in the members of the Raizal family, it was 241 ± 31; and in the two non-family related Raizal participants, it was 210 ±2. The mean number of non-synonymous single nucleotide variants and InDels that have already been reported were 10,053 and 185, respectively. The number of reported non-synonymous single nucleotide variants shared by family members were 33.88; and InDels 68. When comparing the molecular variants of the members of the family vs. non-members, it was found that they didn’t share 423 non-synonymous single nucleotide variants, nor 11 InDels. These possibly pathogenic molecular variants were distributed in 197 genes. By filtering the genes with the list available in DisGeNET, six genes associated with glaucoma were identified.
Table 1 Total of molecular variants identified in the exome analysis of members of a Raizal family from the San Andres and Providencia archipelago with a history of glaucoma.
| Variants \ ID | Members of Raizal Family | N.F-g26F | N.F-g27F | ||||||
|---|---|---|---|---|---|---|---|---|---|
| III-5* | III-7* | III-15* | IV-9* | IV-5** | III-14 | IV-6 | |||
| Total of Molecular Variants | 27,423 | 27,012 | 27,325 | 27,168 | 26,646 | 27,082 | 26,746 | 24,886 | 26,532 |
| Non-synonymous single nucleotide variants | 12,683 | 12,330 | 12,461 | 12,335 | 12,073 | 12,434 | 12,246 | 11,410 | 12,135 |
| Reported | 10,152 | 10,070 | 10,071 | 10,042 | 10,066 | 10,051 | 10,043 | 10,014 | 10,056 |
| Non-reported | 595 | 550 | 586 | 605 | 460 | 561 | 577 | 621 | 623 |
| InDels | 225 | 247 | 248 | 305 | 225 | 216 | 219 | 209 | 212 |
| Reported | 189 | 203 | 190 | 196 | 193 | 187 | 179 | 171 | 187 |
| Non-reported | 36 | 44 | 58 | 109 | 32 | 29 | 40 | 38 | 25 |
*Diagnosed with glaucoma.
**With suspected diagnosis of glaucoma.
SNV: Single nucleotide variant. InDels: insertions and deletions.
Table 2 shows the genes identified with their single nucleotide variant molecular variants that are present in family members, compared with non-members. The identified genes were: BTC (betacellulin); CENPJ (Centromere J protein); DHDDS (dehydrodolichol diphosphate synthase); TFR (Transferrin Receptor); and WLS (receptor for Wnt proteins on secretory cells).
Table 2 Genes and molecular variants identified in members of a Raizal family from the San Andres and Providencia archipelago with a history of glaucoma
| GEN \ ID | Members of the Raizal Familia | N.F-g26F | N.F-g27F | ||||||
|---|---|---|---|---|---|---|---|---|---|
| III-5* | III-7* | III-14 | III-15* | IV-5** | IV-6 | IV-9* | |||
| BTC | rs11938093 | rs11938093 | rs11938093 | rs11938093 | - | - | - | - | - |
| rs28549760 | rs28549760 | rs28549760 | rs28549760 | rs28549760 | rs28549760 | rs28549760 | - | - | |
| CENPJ | - | rs17402892 | - | - | - | - | - | - | - |
| - | rs9511510 | - | - | - | - | - | - | - | |
| - | rs17081389 | - | - | rs17081389 | - | - | - | - | |
| rs7336216 | - | rs7336216 | rs7336216 | - | rs7336216 | - | - | ||
| DHDDS | rs3816539 | rs3816539 | rs3816539 | rs3816539 | rs3816539 | rs3816539 | rs3816539 | - | - |
| TFRC | rs3817672 | rs3817672 | - | rs3817672 | rs3817672 | rs3817672 | rs3817672 | - | - |
| WLS | rs983034 | rs983034 | rs983034 | rs983034 | rs983034 | rs983034 | rs983034 | - | - |
*Diagnosed with glaucoma.
**With suspected diagnosis of glaucoma.
Table 3 presents the damage prediction of each identified non-synonymous single nucleotide variant. The WLS gene presented the non-synonymous single nucleotide variant rs983034 with a prediction of high damage to protein structure and function. The rs983034 was present in all members of the Raizal family; but it was absent in non-family members (Table 2). The TFRC gene presented the non-synonymous single nucleotide variant rs3817672 with a prediction of great damage in six of the members of the Raizal family. In the CENPJ gene, rs7336216 was identified with high damage prediction in four family members. Finally, the BTC gene presented the non-synonymous single nucleotide variant rs11938093 with high damage prediction in four of the family members. The DHDDS gene did not present non-synonymous single nucleotide variants with predicted harmfulness.
Table 3 Non-synonymous single nucleotide variants molecular variants and their damaging potential in genes associated with glaucoma pathology identified in an Afro-Colombian Raizal family from San Andres and Providencia.
| single nucleotide variants ID | Chromosome (CRCh38/hg38) | Gen | Nucleotide change | Amino acid change | CADD value | CADD_MSC value | Damaging potential | Global allele frequency * |
|---|---|---|---|---|---|---|---|---|
| rs11938093 | cr4:74750631 | BTC | A>T | L124M | 24,4 | 3,313 | Alto | 0.199880 |
| rs28549760 | cr4:74794307 | BTC | A>C | C7G | 0,02 | 3,313 | Bajo | 0.250799 |
| rs17081389 | cr13:24912863 | CENPJ | G>C | P55A | 1,513 | 10,02 | Bajo | 0.028155 |
| rs17402892 | cr13:24905403 | CENPJ | A>C | S879A | 0,022 | 10,02 | Bajo | 0.061102 |
| rs7336216 | cr13:24912839 | CENPJ | C>G | D63H | 22,5 | 10,02 | Alto | 0.024760 |
| rs9511510 | cr13:24912773 | CENPJ | G>T, A | P85A, T | 0,032 | 10,02 | Bajo | 0.060903 |
| rs3816539 | cr1:26460136 | DHDDS | G>A | V253M | 16,19 | 24,721 | Bajo | 0.455272 |
| rs3817672 | cr3:196073940 | TFRC | C>T | G142S | 4,709 | 3,313 | Alto | 0.306310 |
| rs983034 | cr1:68137903 | WLS | C>T | V463I | 22,5 | 5,269 | Alto | 0.236621 |
BTC (betacellulin); CENPJ (Centromere J protein); DHDDS (dehydrodolichol diphosphate synthase); TFR (Transferrin Receptor); and WLS (receptor for Wnt proteins on secretory cells). CADD: Combined Annotation Dependent Depletion; MSC: Mutation Significance cutoff. single nucleotide variants in bold have a high damage prediction according to the CADD algorithm. *Taken from the database of 1,000 genomes.
Discussion
Complete exome analysis of a Raizal family group from the archipelago of San Andres and Providencia, Colombia, was performed in the present study. Several members of this family had been diagnosed with glaucoma, and it was possible to identify four harmful non-synonymous single nucleotide variants in genes associated with the disease: rs11938093 in the BTC gene; rs7336216 in the CENPJ gene; rs3817672 in the TFRC gene; and rs983034 in the WLS gene. Interestingly, single nucleotide variant rs983034 in the WLS gene was identified only in members of the Raizal family. Likewise, the non-synonymous single nucleotide variants rs3816539 in the DHDDS gene and rs28549760 in the BTC gene were identified only in family members but with low damage prediction. Based on the heredogram that was constructed with the information obtained from the participants and the clinical history of those diagnosed with glaucoma (Figure 1), it can be suggested that the non-synonymous single nucleotide variants rs983034, rs3816539 and rs28549760 present autosomal dominant segregation, just like the disease. This coincides with Meneses et al., 2011 27, who reported that 43.1% of glaucoma cases are familial; these, 56.3% show an autosomal dominant inheritance pattern, 38.9% an autosomal recessive form, and 2.8% do not present a defined pattern.
According to the clinical history records, the relatives with glaucoma from levels I to III in the hederogram were diagnosed after the age of 55 years, while the three relatives with glaucoma, and the one with suspected level IV glaucoma were diagnosed before the age of 47 years. Due to the above, it is possible that (they / some of them) are presenting a type of juvenile glaucoma caused by low-frequency variants; however, it is necessary to increase the number of samples to make future analyses adjusted for age, as well as for other risk factors, such as systemic arterial hypertension and diabetes mellitus 8,9.
It was also possible to determine that African is the main ancestral genetic component of the group of Raizal participants from the archipelago of San Andres and Providencia in the Colombian Caribbean; followed by the European, possibly as a consequence of the arrival of these human groups to the Americas due to the trafficking of enslaved people and ethnic mixture between the 16th and 19th centuries, due to the expansion of the British empire; and later, the Spanish one 28,29. Likewise, ancestral components of East Asia were found, being in one person the main one, possibly due to the arrival of people at the archipelago from China, between 1839 and 1917, in search of work in the growing fields 30. Hence, the expanded denomination of the term Raizal is both for people with African genetic ancestry and Chinese in the archipelago of San Andres and Providencia. The present investigation confirmed that the Raizal family under study had an African genetic ancestral component (Figure 2). Furthermore, studies conducted in African-Americans have indicated that visual impairment, vision loss, and the diagnosis of angle glaucoma are more frequent in this population than in Europeans / European descendants 31,32.
On the genes identified with non-synonymous single nucleotide variant with the prediction of high protein damage, rs983034 was identified in the WLS gene in all members of the Raizal family (Tables 2 and 3). The WLS gene encodes a receptor for Wnt proteins in secretory cells that regulates this signaling pathway. The Wnt signaling pathway is involved in highly conserved signal transduction during development that regulates various cellular functions 33,34. In eye development, Wnt signaling controls multiple developmental processes and morphogenic patterns. These processes include the dorsoventral pattern of the optic cup, the lens, the retinal pigment epithelium, the vascular system, and the ciliary margin 35-39. Likewise, Wnt signaling has been widely related to ocular diseases such as retinal degeneration, cataracts, congenital ocular dysfunctions 40 and glaucoma 41,42.
The main risk factor for glaucoma, especially for primary open-angle glaucoma, is increased intraocular pressure. Elevated intraocular pressure in patients with glaucoma is due to glaucomatous lesions of the trabecular meshwork and impaired function of the latter, with increased resistance to aqueous humor outflow due to excessive deposition of extracellular matrix proteins. A study showed that the WLS-regulated Wnt cell signaling pathway controls homeostasis of the trabecular meshwork in a spatiotemporal manner; and its inhibition is associated with an increase in intraocular pressure, which leads to the pathology of glaucoma; on the contrary, its activation after inhibition reverses the pathological phenotype 43.
The non-synonymous single nucleotide variant rs3817672 in the TFRC gene with high damage prediction was identified in six of the seven family members. The family member in whom this single nucleotide variant was not identified had a negative diagnosis for glaucoma. The TFRC gene encodes a cell surface receptor required for cellular iron uptake through the process of receptor-mediated endocytosis. The degradation of TFCR is induced by the overexpression of the protein Optineurin (OPTN) by a mechanism that is still unknown 44, and the recruitment of the RAB12 protein, leads to the death of retinal ganglion cells by autophagy 45, which could ultimately lead to the pathology of glaucoma.
Two non-synonymous single nucleotide variants were identified in the BTC gene, where one of them was identified in four members of the family, rs11938093, with a prediction of high damage. The BTC gene encodes a protein part of the epidermal growth factors family. BTC plays a crucial role in the regulation of retinal vascular permeability 46. A study showed that glycosylation of this protein has proliferative effects on retinal pigment epithelial cells, suggesting that it is closely related to retinal vascular etiology and pathogenesis 47. Furthermore, a study conducted in a mouse animal model with glaucoma showed that BTN messenger RNAs are inhibited by microRNA-149, which was related to increased apoptosis of murine retinal ganglion cells 48.
The non-synonymous single nucleotide variant rs7336216 predicting high damage in the CENPJ gene was identified in four of the seven members of the raizal family. CENPJ encodes a protein that belongs to the family of centromere proteins, which, during cell division, play an important role in maintaining the integrity of the centrosome. Likewise, it has been described that the CENPJ protein can function as a transcriptional coactivator in the Stat5 signaling pathway, and NF-kappa B. single nucleotide variant in this gene have been related to diseases such as primary autosomal recessive microcephaly and Seckel syndrome 49. Seckel syndrome is associated with ocular defects in humans, including spontaneous lens dislocation, myopia, astigmatism, and retinal degeneration 50. Low expression of CENPJ has been detected in the retinal neuroblast layer of mice that have exhibited a number of ocular abnormalities compared to healthy mice 51.
Regarding the DHDDS gene, a non-synonymous single nucleotide variant was identified in all members of the Raizal family, rs3816539; however, the prediction of damage was low. The DHDSS gene encodes part of the enzymatic complex dehydrodolichol diphosphate synthase, an enzyme that participates in the synthesis of Dolichol monophosphate at the retina level; dolichol acts as an "anchor" at the level of the membrane of the endoplasmic reticulum of the rods of the eye, allowing the glycosylation of synthesized rhodopsin. The activity of DHDDS must be balanced since the lack of glycosylation of rhodopsin affects both the function of this protein and its intracellular traffic, leading to the progressive degeneration of photoreceptors 52.
Conclusion
In the present study, four non-synonymous single nucleotide variant molecular variants were identified with a prediction of high damage to gene protein structure and function associated with glaucoma pathology in an Afro-Colombian Raizal family with a history of this disease. Interestingly, single nucleotide variant rs983034 in the WLS gene was identified only in members of the Raizal family. Likewise, the non-synonymous single nucleotide variants rs3816539 in the DHDDS gene and rs28549760 in the BTC gene were identified only in family members, but with low damage prediction. Despite the above, we do not rule out the possibility that these last two single nucleotide variants can help screen for the risk opportunity of developing glaucoma in this Raizal family; and in the population with African ancestral components in general.
Due to the small size of the control group sample, there is a limitation in identifying the disease-causing variants in these patients. Thus, given the high frequency in different populations of the selected variants, there is a high probability that these variants were chosen at random because they were not represented in the two control subjects used in the study. Additional studies will be needed to test this hypothesis.











 text in
text in 



