Remark
| 1) Why was this study conducted? |
| This study was conducted from the need for effective extraction and analysis of tumor characteristics from oncology reports recorded in the such registry. |
| 2) What were the most relevant results of the study? |
| An algorithm using artificial intelligence to process natural language was developed. As a result, an adequate concordance with human evaluation about the most critical parameters in determining tumor frequencies, topography, and morphology was achieved. |
| 3) What do these results contribute? |
| This study presents a tool for classifying oncological diseases and a notification system that facilitates the implementation of a cancer registry. |
Introduction
Cancer registries collect, store, analyze, and access cancer data of a given population 1. They record patient demographics, cancer characteristics, treatment information, and patient outcomes to monitor and identify cancer prevention and control methods. Information comes from healthcare databases, including electronic health records, diagnostics imaging, laboratory tests, and pathology reports which result in structured variables and unstructured data 2. Usually, the most relevant information for cancer cases is included in the pathology report. Those reports follow a pre-established format in an unstructured text that is ungrammatical, fragmented, and abbreviated, with linguistic variability amongst pathologists 3. In this scenario, the extraction task requires a time-consuming and laborious effort that humans manually perform.
Natural Language Processing is a subfield of artificial intelligence that combines linguistic, statistical, and computational techniques to analyze and represent human language in a machine-readable format 4. Natural Language Processing has demonstrated the potential to automatize healthcare information extraction processes 5,6.
Studies using Natural Language Processing applications to extract information from cancer pathology reports have been published in English, Dutch, French, German 7, and Italian 8 and are mainly focused on extracting a single characteristic 8 or a few of them 9. A similar effort has been performed regarding data extractions from radiological 10 and public health reports in Spanish 11. Using additional techniques like deep learning, another artificial intelligence subfield, researchers have extracted features from lung cancer-free text in clinical records in Spanish 12. To do this, they follow a three-step process that includes using Natural Language Processing for name entity recognition. However, their model uses supervised learning techniques (i.e., deep learning), requiring the manual annotation of seven features (cancer entity, stage, dates, events, family members, treatment and drug) in 14,759 sentences. Further refinements 13, using deep learning techniques and annotated texts, are pursued by the authors to extract eleven similar features.
In this project, we aimed to implement an algorithm to automatically extract 20 key cancer characteristics in oncology pathology reports written in Spanish from a hospital-based cancer registry.
Material and Methods
Dataset
Fundación Valle del Lili is a non-profit, highly complex University Hospital in Cali, Colombia; its hospital-based cancer registry includes patients diagnosed with cancer from January 1 2014 to November 13 2019 (14. Data are stored in a computer platform owned by the institution, which meets the 2016 Facility Oncology Registry Data Standards (FORDS) recommendations 15.
We obtained a text corpus of cancer pathology reports from the hospital-based cancer registry. The corpus consisted of unstructured text from 22322 anonymized pathology reports of cancer cases diagnosed from January 1, 2014, to November 13, 2019. Each report included three sections as free-text: pathology diagnosis, macroscopic and microscopic description (Table 1).
Table 1 Spanish Pathology report examples in free text.
| Macroscopic description | Microscopic description | Diagnosis |
|---|---|---|
| Tres fragmentos de mucosa gástrica. Se procesa todo en 1 canastilla | Mucosa gástrica antral infiltrada por glándulas malignas | Mucosa gástrica antral. Biopsia Adenocarcinoma bien diferenciado |
| Se recibe en un tubo con EDTA, aproximadamente 4 ml de médula ósea | Población patológica: 48% de blastos mieloides CD34+, CD117+, CD33+, CD13+, cMPOdim, CD56 parcial, HLA-DR+. | Proliferación de blastos mieloides del 48% compatibles con leucemia mieloide aguda con cambios relacionados a mielodisplasia |
| Se recibe rotulado como “dorso lumbar izquierda”, fragmento de piel de 5.5x4.5x3.0 cm | Melanoma nodular fase de crecimiento vertical Nivel de Clark IV Espesor de Breslow 1.5 cm | Dorso lumbar izquierdo. Lesión. Biopsia: Los hallazgos histológicos observados muestran melanoma nodular |
| “mama derecha” se reciben 11 fragmentos de tejido, el mayor de 1.6x0.2cm. Se procesa todo en 3 canastillas. | 5. Patrón morfológico y tipo histológico: Carcinoma invasivo, tipo indeterminado | Mama derecha. Biopsia Trucut: Carcinoma invasivo, tipo indeterminado score de Nottingham 3 (9/9) |
| “Tumor colon derecho”: siete fragmentos de tejido blanquecino y blando, el mayor de 0.2x0.2 cm. Se procesa todo en una canastilla. | La totalidad de la muestra corresponde a una lesión neoplásica maligna de origen epitelial | Mucosa de colon. Colonoscopia. Lesión. Biopsia: Adenocarcinoma |
Descriptors to extract from pathology reports
Twenty cancer essential characteristics were extracted from oncology pathology reports embedded in the hospital-based cancer registry. These descriptors of interest were included in the "Cancer identification" module. In addition, the recommendations of the 2016 Facility Oncology Registry Data Standards15 were adapted to the Mandatory Notification Record established by the Instituto Nacional de Salud of Colombia 16 in the 247 Resolution of 2014.
We divide each extracted descriptor into four groups according to its clinical relevance and the kind of values they could take.
Primary descriptors
Topographic (which identifies the anatomical site where the malignancy was found), and morphologic (which determines the microscopic type of the tumor cells) variables contain the most relevant information in the pathology report as they constitute the base of case classification. Both descriptors take values in the form of free text.
Complementary descriptors
These descriptors contain valuable information concerning the primary tumor identified with the main descriptors. They can be classified into different categories, as shown in Table 2.
Table 2 Descriptors extracted from each oncology pathology report. The first column shows the descriptor name and defintion, the second the type of values it can take, and the third a description of these values.
| Descriptor name and definition | Value | Meaning | |
|---|---|---|---|
| Main descriptors | Topography: Identifies the anatomical site where malignancy was found | Free text | As Pathologist wrote |
| Morphology: Identifies the microscopic type of tumor cells | Free text | As Pathologist wrote | |
| Complementary descriptors | Laterality: Identifies the side of a paired organ or the body side on which the tumor originated | 0 | Non paired organ |
| 1 | Right side | ||
| 2 | Left side | ||
| 9 | Paired organ, unknown side | ||
| Behavior: Describes the tumor's clinical behavior | 0 | Benign | |
| 1 | Borderline | ||
| 2 | In situ | ||
| 3 | Invasive | ||
| Grade: Describes the tumor's resemblance to normal tissue | 1 | Well-differentiated | |
| 2 | Moderately differentiated | ||
| 3 | Poorly differentiated | ||
| 4 | Undifferentiated | ||
| 5 | T cells | ||
| 6 | B cells | ||
| 8 | NK cells | ||
| 9 | Unknown | ||
| Method of Assessment for Solid Tumors: Records the diagnostic method used to diagnose solid cancer | 0 | Not a solid tumor | |
| 1 | Positive histology | ||
| 2 | Positive cytology | ||
| 9 | Unknown | ||
| Method of Assessment for hematological Tumors: Records the diagnostic method used to diagnose hematological cancer | 0 | Not a hematological tumor | |
| 3 | Positive histology plus | ||
| Diagnostic Procedure: Records the diagnostic procedure performed to confirm cancer | 1 | The biopsy is not the primary site | |
| 2 | Biopsy primary site | ||
| 3 | Exploration | ||
| 5 | Surgery | ||
| 9 | Unknown | ||
| Lymphovascular Invasion: Indicates the presence or absence of tumor cells in lymphatic channels or blood vessels | 0 | Absent | |
| 1 | Present | ||
| 8 | Nonapplicable | ||
| 9 | Unknown | ||
| Surgical Margins: Records if the tumor margins presented the macroscopic or microscopic compromise | 0 | Without residual tumor | |
| 1 | With residual tumor; NOS | ||
| 2 | Microscopic residual tumor | ||
| 3 | Macroscopic residual tumor | ||
| 9 | Unknown | ||
| Liver metastasis: Identifies whether the liver is an involved metastatic site | 0 | Absent | |
| 1 | Present | ||
| 8 | Nonapplicable | ||
| 9 | Unknown | ||
| Lung metastasis: Identifies whether the lung is an involved metastatic site | 0 | Absent | |
| 1 | Present | ||
| 8 | Nonapplicable | ||
| 9 | Unknown | ||
| Brain metastasis: Identifies whether the brain is an involved metastatic site | 0 | Absent | |
| 1 | Present | ||
| 8 | Nonapplicable | ||
| 9 | Unknown | ||
| Bone metastasis: Identifies whether the bone is an involved metastatic site | 0 | Absent | |
| 1 | Present | ||
| 8 | Nonapplicable | ||
| 9 | Unknown | ||
| Distant lymphatic nodes metastasis: Identifies whether any distant lymphatic nodes are found to contain metastasis | 0 | Absent | |
| 1 | Present | ||
| 8 | Nonapplicable | ||
| 9 | Unknown | ||
| Other metastasis: Identifies whether a different anatomical region to the liver, lung, brain, bone and distant lymphatic nodes is an involved metastatic site | 0 | Absent | |
| 1 | Present | ||
| 8 | Nonapplicable | ||
| 9 | Unknown | ||
| Special Descriptors | TNM: Records the TNM stratification registered by the Pathologist | Free text | As Pathologist wrote |
| Tumor size: Records the most accurate measurement of a solid primary tumor | Numeric | Two or three dimensions | |
| Examined lymphatic nodes: Records the exact number of regional lymph nodes examined by the Pathologist | Numeric | Numeric | |
| Positive lymphatic nodes: Records the exact number of regional lymph nodes examined by the Pathologist and found to contain cancer | Numeric | Numeric |
Metastasis-related descriptors
These descriptors identify if the mentioned organ is a metastatic site and evaluate the pulmonary, bone, liver, brain, and distant lymph nodes compromise, as well as other metastasis. Descriptors scoring was: 0: NOT a metastatic site, 1: it is a metastatic site, 8: nonapplicable, 9: unknown.
Special descriptors
The descriptors in this group have different possible values and provide complementary information that might not be present or even applicable in many pathology reports. These descriptors are: number of lymphatic nodes examined, number of positive for malignancy lymphatic nodes sectioned near the tumor, the tumor size and the tumor, lymphatic nodes, and metastasis (TNM)-based staging.
Each descriptor could take up to two values: nonapplicable (NA) and unknown or unreported (NR). Nonapplicable was used when the descriptor did not apply to the procedure or cancer type reported; for example, it does not make sense to assess the residual tumor and surgical margins in the case of a biopsy. Unreported was used when the descriptor applied to the case but was not mentioned in the report.
Construction of the algorithm
Descriptors from the pathology report text were extracted using Natural Language Processing techniques, particularly the matching of regular expressions and the fuzzy matching of strings.
This project was developed in Python, and a module containing an algorithm for extracting each descriptor was implemented. Each algorithm loosely obeyed the following steps (Figure 1):
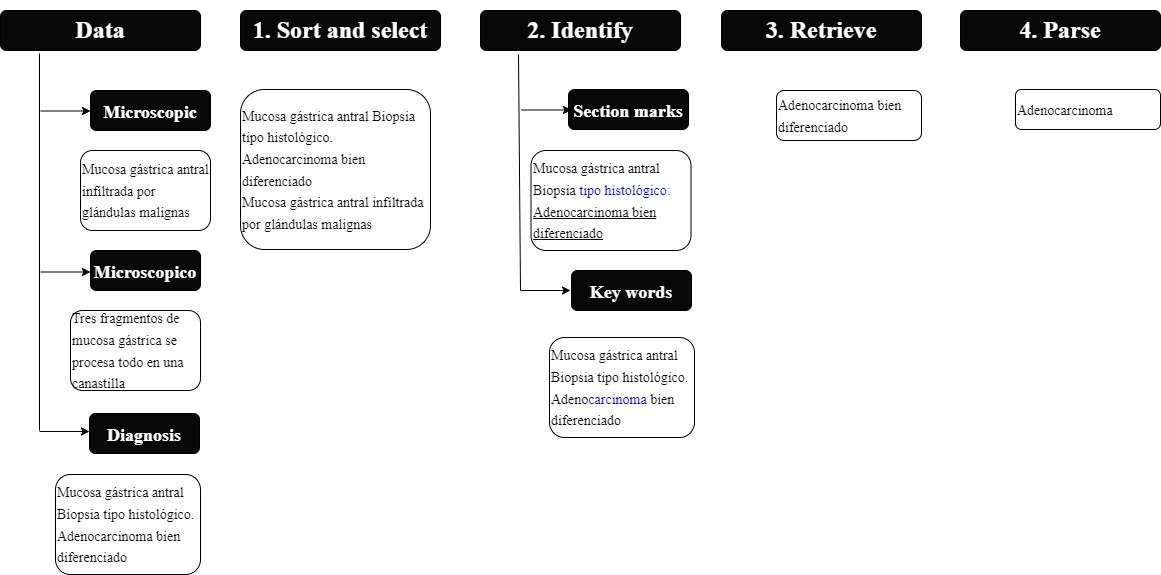
Figure 1 Algorithm: the figure shows the process followed to identify and retrieve the relevant characteristics of the oncology pathology report. The algorithm is feed with three types of data: microscopic, macroscopic and diagnosis data. It then follows a four step process in which the data is sorted (step 1), characteristics are identified inside the text (step 2) and finally, they are retrieved (step 3) and parsed or tokenized into grammatical parts (step 4).
1. Choice of pathology sections and their order for description search.
2. Identify the marker that introduced the value of the descriptor (in case it was explicitly stated). For instance, the tumor size was usually preceded by the phrase "Tamaño del tumor".
3. Identifying keywords directly related to the descriptor in case the value was tacitly mentioned in the text.
4. Extraction of relevant text.
5. Analyzation of the value of the such text.
The following paragraphs describe the algorithms for each kind of descriptor in more detail.
Primary descriptors: Topography and morphology
For each variable, a thesaurus was built based on the corresponding section of the International Classification of Diseases for Oncology (ICD-O) Spanish translation 17. This thesaurus identified the main keywords in every topography and morphology category. Those keywords (e.g., "carcinoma") were searched in the diagnosis section of the pathology report text first, followed by other sections. Once a match was found, a secondary search for relevant modifiers for the keyword (e.g. " ductal", "papillary" etc.) was performed in nearby words.
Complementary descriptors
This group of descriptors offered complementary information on the performed examination and the results found. All were calculated after topography and morphology were determined. Each descriptor had a few possible values, depending on whether the cancer was established as a solid tumor or a hematologic malignancy (such distinction can be made based on topography or morphology).
Laterality was implemented as a lateral topography computation, first by verifying if the organ was paired and then its side among the modifiers found. Behavior was found in the diagnosis section, usually close to the morphology and in some cases, implied by it. Due to the data nature, the predetermined value was malignancy when was not explicitly stated.
The Grade was determined from three possible sources: 1. A keyword for differentiation explicitly stated or near the morphology declaration, for instance: well-differentiated (i.e., "bien diferenciado"). 2. A global grade number or a numerical score for a specified set of topographies. For instance, Nottingham scored in breast cancer. 3. For hematological malignancies, the kind of lymphocyte involved was either explicitly stated or derived from a biological marker.
The assessment method and the Diagnostical procedure substantially depended on the distinction between solid and hematologic. The examination type complements this information, and the keywords search among microscopic or macroscopic descriptions.
The examination of residual tumor and surgical margins only proceeded when a surgical procedure was performed and was specified as micro or macro depending on the residual tumor size. When evaluated, the presence or absence of lymphovascular invasion was usually explicitly stated in the microscopic description.
Metastases-related descriptors
Six descriptors study the spread of cancer according to compromised organs. These were calculated simultaneously following a two-step procedure: first, identification of each metastasis mentioned in the report and extraction from the surrounding texts. Then, a mention for each specified organ was searched in the texts; if no organ was found but metastases were mentioned in a non-negative manner, these were classified as "other Metastases".
Two special conditions were taken into account in this algorithm: first, exclusion of cancer in the primary organ as a possible metastatic site, and second, differentiation between regional and distant lymphatic nodes.
Especial descriptors
These were determined based on the applicability rules of the descriptor and some manipulation of numbers reported. Finally, the TNM staging was extracted by a global search based on regular expressions, considering repetition and scares code statements.
For instance, the TNM code could be distributed in a paragraph first indicating the T value and a couple of sentences after stating the N and M values.
The tumor size was searched solely when the resection was performed. For extraction, the context of every number that resembled a measure (e.g. "1.2 cm") was inspected to establish if the tumor was mentioned. The number of lymphatic nodes examined and positive nodes were calculated from a context inspection of the numbers present in the diagnosis or in the microscopic description of the pathology.
Algorithm evaluation
During the algorithm's development, a team of experts in our institution selected a subset of pathology reports and executed a manual extraction of descriptors for such reports. This human team included a general physician, a pathologist, and an hemato-oncologist. Reports for manual extraction were carefully chosen to ensure the inclusion of a wide range of pathology reports. Special attention was given to including representatives of every database, most of the common cancer types and stages, and every kind of procedure.
In order to assess and improve the algorithm's performance, the manual and algorithmic information extraction was compared in three incremental cycles (first 20 reports, then 42, and finally 140). After each evaluation cycle, possible algorithm error sources were identified, and many suggestions for improvement were made and implemented.
The metrics used for measuring the algorithm's performance depended on the kind of values that each descriptor could take:
The values were considered free text for the main primary descriptors, and a fuzzy matching score was calculated. This score is based on the Levenshtein distance between the text extracted by the algorithm and the human team; this distance measures the number of edits (adding, erasing, or replacing a character) needed to transform a word into another. The distance is scaled to obtain a score that ranges from 0 to 100. Therefore a score of 100 means that the words in both texts are identical, and a score of 0 means that both texts have no characters in common.
The values were split into a small number of classes for the other descriptors. Hence we used four common metrics for a multiclass classification problem: the overall accuracy and the macro averaged precision, recall, and f-score.
The overall accuracy measures the fraction of reports correctly classified among all reports, where correctly means that human and algorithmic extraction coincide.
For each possible value of the descriptor, we compute the precision, recall and f-score in a one versus the rest strategy according to the next formulae:
The precision measures how good the algorithm is at differentiating this class from the others, and the recall measures how good the algorithm is at capturing all the instances of the same class. Since those two objectives are complementary, the f-score is a compromise between both.
Finally, the arithmetic mean of each metric is taken over all the possible values of a descriptor. This is known as the macro average.
In addition, for the special descriptors, where the nonapplicable or non-reported values represent a significant proportion, a categorical analysis was performed between reported, non-reported, and nonapplicable classes before proceeding to the analysis of the reported values.
In order to perform a larger-scale validation of the algorithm, a website (available at one of our institutional computer platforms) for the algorithm was developed, with open access to all interested external users who may voluntarily participate in its evaluation and improvement (https://oncologia-web-app-dev.uc.r.appspot.com/polls/).
Results
This section summarises the comparison between the human and algorithmic descriptors extraction for the pathology reports chosen for validation. The evaluation was performed as described in the previous section.
Primary descriptors
The validation was performed in 140 pathological reports. Topography was identified by both the human and the algorithm in all reports. The human identified morphology in 138 reports and by the algorithm in 137.
A fuzzy matching score was calculated between the values on the reports where both the human and algorithm extracted the descriptor. Table 3 summarizes the distribution of that score calculated for each descriptor. Notice that the matching score is above 90.0 for three-quarters of cases in the Morphology text.
Table 3 Summary statistics for the fuzzy matching score between human and algorithmic extraction of free text descriptors. The table displays the number of reports validated and the mean, standard deviation and quartiles of the score.
| Descriptor | Count | Mean | std | Min | 25% | 50% | 75% | Max |
|---|---|---|---|---|---|---|---|---|
| Topography | 140 | 68.27 | 25.22 | 0.0 | 45.0 | 77.0 | 90.0 | 100.0 |
| Morphology | 137 | 89.45 | 10.64 | 31.0 | 90.0 | 90.0 | 95.0 | 100.0 |
Complementary descriptors
Precision, recall, and f-score were calculated for each possible value of the descriptor and then averaged. The overall precision corresponds to the fraction of reports where the manual and algorithmic extraction for the descriptor match. Table 4 summarises the algorithm's precision, recall, and accuracy for each categorical descriptor in the validation subset of 42 reports.
Table 4 Performance measures of the extraction algorithm when applied to categorical characteristics. Precision measures the number of correctly classified reports among the total number of reports assigned to the class by the algorithm. Recall, measures the number of reports correctly classified among the number of true (i.e., human classified) reports in that class. The f-score is the harmonic mean of precision and recall. For multiclass characteristics precision, recall and f-score are averaged over classes (macro average). Overall accuracy is the number of reports correctly classified among the total number of reports evaluated.
| Descriptor | Macro Precision (%) | Macro Recall (%) | Macro f-score (%) | Overall Accuracy % (n/N) | |
|---|---|---|---|---|---|
| Complementary descriptors | Laterality | 66.2 | 50.0 | 52.9 | 64.3 (27/42) |
| Behavior | 57.1 | 92.7 | 58.6 | 85.7 (36/42) | |
| Grade | 70.3 | 64.8 | 79.6 | 76.2 (32/42) | |
| Method of Assessment for Solid Tumors | 78.6 | 94.8 | 78.4 | 85.7 (36/42) | |
| Method of Assessment for Hematological Tumors | 100 | 100 | 100 | 100 (42/42) | |
| Diagnostic Procedure | 95.0 | 83.7 | 87.2 | 90.5 (38/42) | |
| Lymphovascular Invasion | 82.5 | 91.2 | 83.9 | 85.7 (36/42) | |
| Surgical Margins | 94.4 | 77.2 | 82.8 | 90.5 (38/42) | |
| Pulmonary Metastasis | 100 | 100 | 100 | 100 (42/42) | |
| Osseous Metastasis | 92.8 | 50.0 | 96.3 | 92.9 (39/42) | |
| Hepatic Metastasis | 75.0 | 66.7 | 83.3 | 97.6 (41/42) | |
| Brain Metastasis | 50.0 | 50.0 | 100 | 97.6 (41/42) | |
| Distant Lymph Nodes Metastasis | 50.0% | 97.6 | 98.8 | 97.6 (41/42) | |
| Other Metastasis | 98.8 | 75.0 | 82.7 | 97.6 (41/42) | |
| Special descriptors | Examined Regional Nodes | 92.3 | 100 | 96.0 | 41.7 (5/12) |
| Positive Regional Nodes | 92.3 | 100 | 96.0 | 58.3 (7/12) | |
| Tumor Size | 85.7 | 75.0 | 80.0 | 50.0 (6/12) | |
| TNM-based Staging | 100 | 75.0 | 85.7 | 100 (3/3) |
Special descriptors
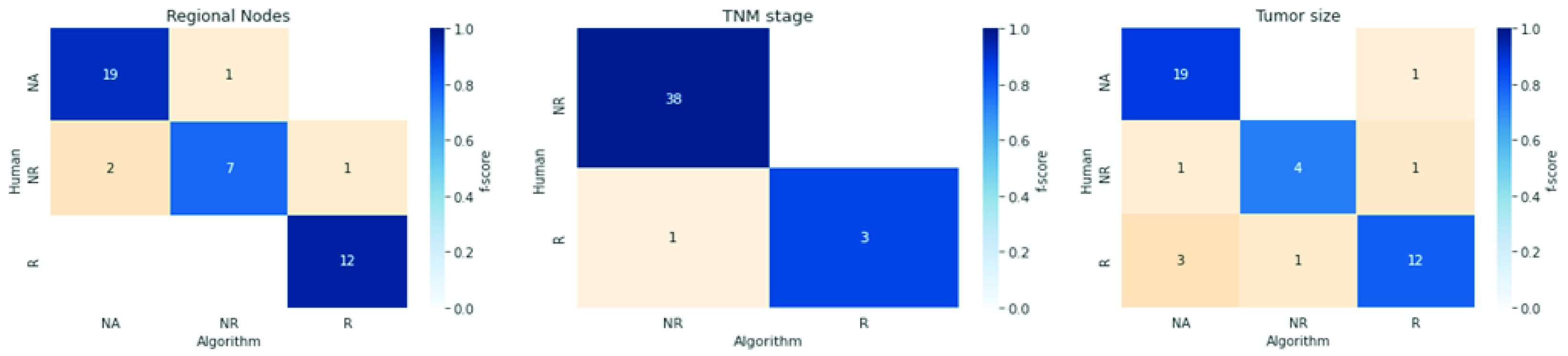
The analysis is executed in two steps for these descriptors. First, we measured the algorithm performance in differentiating reported values from unreported and nonapplicable. Afterward, we measured the precision of the reported values. Figure 2 shows confusion matrices colored by f-score for each descriptor.
Discussion
Here we present an effort to combine Natural Language Processing developments with human effort to optimize the information extraction results for the tumor module of our hospital-based cancer registry.
Data extraction using a regular-expressions approach can extract multiple specimen attributes from free-text pathology reports in Spanish with acceptable accuracy and precision. In cases selected for validation, the average fuzzy matching score was 68.3 for topography and 89.5 for morphology. Complementary descriptors presented precision and recall between 50% and 100% and F-score between 52.9% and 100%. Among the reported cases, precision ranged from 92.3% to 100%, recall from 75% to 100%, and F-score between 80% and 96%.
These developments could assist in accurately extracting information from hospital cancer registries that face the challenge of handling enormous volumes of information. Based on the algorithmic extraction of descriptors, statistical analyses of these pathology reports are now feasible.
Although the precision of the model is high, other metrics, such as recall, show there's room for improvement. The recall shows that the rules created via regular expressions were not enough to capture a significant number of characteristics of the tumors. This may be caused by underlying language patterns that doctors are unaware of because (1) they are infrequent or (2) they may be too complex to identify. These two obstacles seem insuperable using regular expressions and fuzzy matching of strings since all the potential cases would have to be included, many of which are unknown by pathologists. This is the most critical limitation of regular expressions.
Nevertheless, other methodologies in Natural Language Processing could prove to be more accurate in these cases; this tool is an approach that could significantly increase the recall of this application via machine learning models. Machine learning models in text data can identify underlying patterns that humans cannot, overcoming the limitations of prior knowledge constraints. Deep learning methodologies, such as recurrent neural networks, word2vec, and transformers, can capture the meaning of words/terms from their context, understanding context as the language around them. With enough data, these models could leverage information in the text, such as longevity, location within the text, and order of occurrence, to deduce complex correlations and extract the characteristics more accurately. Further research would go in this direction, where learning models are trained to overcome the limitations caused by complexity or infrequent linguistic patterns.
Applying the algorithm in pathology reports with unstructured or structured texts may aid institutions in hospital cancer registry implementation. The extracted data will allow a tumor (ICD-O-M) classification according to location, size, lymphovascular involvement, lymph node compromise, metastasis, and determining staging with TNM.
Limitations to the algorithm include the human supervision required for data extraction. The algorithm improves when essential malignancy data is recorded in pathology reports with cancer protocol templates. Further studies are needed to demonstrate the algorithm reach in a larger corpus of information.











 texto em
texto em