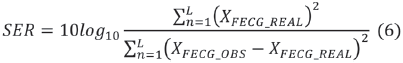Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Iteckne
Print version ISSN 1692-1798
Iteckne vol.11 no.2 Bucaramanga July/Dec. 2014
Fetal electrocardiogram extraction using hybrid BSS technique: COMBI and MULTICOMBI algorithms
Extracción del electrocardiograma fetal usando una técnica hibrida basada en BSS: Algoritmos COMBI y MULTICOMBI
Luis Omar Sarmiento-Álvarez1, José Millet-Roig2, Alberto González-Salvador3
1 Ph.D (c), Universidad Santo Tomás. Bucaramanga, Colombia. luisomar.sarmiento@gmail.com.
2 Ph.D Universidad Politécnica de Valencia. Valencia, España. jmillet@ jmillet@eln.upv.es.
3 Ph.D Universidad Politécnica de Valencia. Valencia, España. agonzal@dcom.upv.es.
ABSTRACT
In this paper, we use two algorithms for obtaining fetal ECG from abdominal ECG. The algorithms are MULTICOMBI and COMBI, which are a combination of EFICA and WASOBI algorithms. The performance of the algorithms COMBI, MULTICOMBI, WASOBI, EFICA and traditional JADE algorithm are compared. A semi synthetic database and two actual databases are used to compare the performance of algorithms using as parameter the signal to error ratio SER. It is found that the COMBI and MULTICOMBI algorithms show better performance than the JADE, EFICA and WASOBI algorithms.
KEYWORDS: abdominal ECG, COMBI, fetal ECG, MULTICOMBI, SER, SIR.
RESUMEN
En este artículo se emplean dos algoritmos para obtener el ECG fetal a partir del ECG abdominal. Los algoritmos son MULTICOMBI and COMBI, los cuales son una combinación de los algoritmos EFICA y WASOBI. SE compara el desempeño de los algoritmos COMBI, MULTICOMBI, WASOBI, EFICA y el tradicional algoritmo de JADE. Para comparar el desempeño de los algoritmos se usa una base de datos semi-sintética y dos bases de datos reales usando como parámetro la relación señal a error SER. Se encuentra que los algoritmos COMBI y MULTICOMBI muestran mejor desempeño que los algoritmos JADE, EFICA y WASOBI.
PALABRAS CLAVE: abdominal ECG, COMBI, fetal ECG, MULTICOMBI, SER, SIR.
1. INTRODUCTION
The analysis of fetal cardiac electrical activity from the fetal electrocardiogram (FECG) provides clinically relevant information on the state of health of the fetus, since it allows detection of fetal abnormalities such as congenital heart block and fetal arrhythmias [1]. The FECG can be obtained from abdominal ECG (AECG) placing electrodes on the mother's abdomen and applying digital signal processing techniques.
The AECG is formed of a mixture of biomedical signals including FECG, maternal ECG (MECG) and other electrical signals generated by the mother and by external electrical interference. The challenge is to extract the FECG from the composite signal, considering that the two signals are superimposed spectrally, and that the fetal QRS complex amplitude may be less than 3% from the maternal QRS complex [2].
To achieve separation of fetal and maternal electrocardiograms are used basically three groups of techniques: adaptive filtering [3], [4], [5], [6], linear decompositions such as Wavelets [7], singular value decomposition (SVD) [8], and blind source separation (BSS) [9], and nonlinear transformations such as techniques based on state space [10] BSS is the most widely used technique for extracting the FECG, since it has been shown that BSS methods are better than the adaptive filters [11], and nonlinear transformations have greater computational load and require some parameters to be set empirically.
In this paper we introduce COMBI[12] and MULTICOMBI [13] algorithms based on hybrid BSS for extracting FECG from the AECG. We compare the performance of COMBI y MULTICOMBI algorithms with the performance of the algorithms JADE [14], WASOBI [15] and EFICA [16] using a semi-synthetic database.
2. METHODS
2.1. Blind Source Separation
BSS consists in recovering a group of signals or sources that have not been directly observed from another group of signals or observations obtained as mixtures of these sources. If the mixture is linear and without noise, we have , where are the observations, the sources and A is the mixing matrix containing the coefficients representing the linear transformation between sources and observations. The BSS problem is solved by finding a linear transformation that complies with s = Wx and W = A-1.
To solve this problem, different methods have been proposed mainly based on Principal Component Analysis (PCA) and Independent Component Analysis (ICA). The underlying difference between PCA and ICA is in the assumptions that underlie the probability distribution functions of the signals contained in x. PCA assumed that x has a Gaussian distribution function that is completely determined by its second-order statistics, while ICA assumes that the x are statistically independent and are not Gaussian and that the distribution function is determined by its higher order statistics.
One way to implement PCA is the Singular Value Decomposition (SVD) of the form x = USVT where VT is the matrix of eigenvalues, so that W = VT. In this case stands SOBI algorithm (Second Order Blind Identification)[17] and its improved version WASOBI (Weights-Adjusted Second Order Blind Identification)[15].
The goal of ICA is to obtain an estimate  mixing matrix A, to calculate its inverse matrix W and thus estimate the sources by s = Wx. ICA requires measuring the Gaussianity of the sources by means of contrast functions as the kurtosis, negentropy or some approximation of the latter that allows reducing the computational cost required for its calculation. In this line stands the FastICA algorithm [18] and EFICA [16]. ICA-based methods are the most used for the analysis of AECG [19], [20] because it is considered that the sources are predominantly non-Gaussian and statistically independent of one another.
Since real-world signals are both Gaussian and non-Gaussian components, it is interesting to test methods that incorporate both approaches. In that sense, algorithms have been developed which combine both WASOBI and EFICA algorithms. These algorithms are called COMBI and MULTICOMBI. In the context of biological signals such algorithms have been applied to EEG signals[21] and fMRI signals [22], but to date it is not known that these algorithms have been applied in the task of extracting the FECG from the AECG.
2.2. Combi y Multicombi algorithms
Under certain conditions, WASOBI and EFICA are asymptotically optimal. WASOBI only take advantage of time-structure, disregarding the statistical distributions of the sources, whereas EFICA can only take advantage of non-Gaussianity of the sources, ignoring any time-structure. However, realistic mixtures are many times compound of sources which present both diverse time-structure and non-Gaussianity, rendering WASOBI and EFICA severely suboptimal. Algorithms COMBI and MULTI-COMBI offer novel schemes for combining WASOBI and EFICA, enabling exploit the strengths of both techniques.
To verify a good degree of separation, is defined G = WA as the gain matrix. For a perfectly estimated de-mixing matrix, W, G is equal to its identity matrix. The performance of blindsource separation algorithms is usually measured by the interference over signal ratio matrix, ISRkl = G2kl/G2kk, k, l = 1, 2, ..., d, where k and l denote the observed and estimated sources, and d is sources number. However, the original mixing matrix, , is not generally known for real data sets.
EFICA requires a user-defined choice of a set of nonlinear functions gk(.), for extracting each of the sources. Then, ISR matrix for the EFICA algorithm can be approximated by:
where γk = βk - µ2k, µk = E[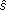 kgk(
kgk(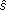 k)], τk = |µk - ρk| ρk = E[g'k(
k)], τk = |µk - ρk| ρk = E[g'k(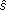 k)], and βk = E[g2k(
k)], and βk = E[g2k(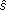 k)] E[.] denotes the expectation operator and g'k(.) denotes the derivative of gk(.), and
k)] E[.] denotes the expectation operator and g'k(.) denotes the derivative of gk(.), and 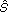 k is the kth observed signals of s [13].
k is the kth observed signals of s [13].
WASOBI is based on approximate joint diagonalization of several (say M) time-lagged estimated correlation matrices, 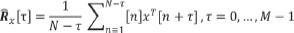 , where x[n] denotes the nth column of x. If all sources are Gaussian AR of order M-1, then under asymptotic conditions the ISR matrix is:
, where x[n] denotes the nth column of x. If all sources are Gaussian AR of order M-1, then under asymptotic conditions the ISR matrix is:
where 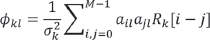 , k and l denote the observed and the estimated sources, σ2k is the variance of the innovation sequence of the kth source,
, k and l denote the observed and the estimated sources, σ2k is the variance of the innovation sequence of the kth source, 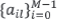 are the auto-regression coefficients of the lth source, and Rk[m] is the autocorrelation of the kth source at time lag m [12].
are the auto-regression coefficients of the lth source, and Rk[m] is the autocorrelation of the kth source at time lag m [12].
In COMBI, the ISR matrices are obviously unknown (nor can they be consistently estimated from the data). However, it is possible to substitute these with the mean ISR, 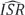 WA and
WA and 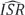 EF. COMBI apply both EFICA and WASOBI to x and estimate
EF. COMBI apply both EFICA and WASOBI to x and estimate 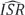 WA and
WA and 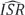 EF select for each source the reconstructed version that has the best total ISR of the two. This basic selection approach can then be turned into a successive scheme, such that in each iteration only the "best" separated sources are "accepted," and the remaining signals (which are still weakly separated mixtures of the remaining sources) are subjected to an additional iteration of separation and selection [12].
EF select for each source the reconstructed version that has the best total ISR of the two. This basic selection approach can then be turned into a successive scheme, such that in each iteration only the "best" separated sources are "accepted," and the remaining signals (which are still weakly separated mixtures of the remaining sources) are subjected to an additional iteration of separation and selection [12].
MULTICOMBI uses a clustering technique based on "multidimensional component". A multidimensional component is a cluster of signal components that can together be well separated from the other components in the mixture, yet are difficult to separate from one another. For EFICA, only components that have (nearly) Gaussian distributions might form such a cluster, hence at most one such cluster may exist. For WASOBI, any components sharing similar correlation structures (i.e., power spectra) are hardly separable from one another, but may be easily separated as a cluster, hence several such clusters might coexist [13]. MULTICOMBI uses this clustering technique in which both algorithms, EFICA and WASOBI, are run on the set of unseparated sources 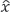 and their
and their 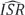 EF and
EF and 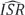 WA, in (1) and (2), are estimated. The signals are then clustered depending on whether their specific ISRkl is lower for the EFICA or WASOBI case. Then, the process is repeated until all clusters are singletons, ie. only contain one signal per cluster, and the signals are hence optimally separated [23].
WA, in (1) and (2), are estimated. The signals are then clustered depending on whether their specific ISRkl is lower for the EFICA or WASOBI case. Then, the process is repeated until all clusters are singletons, ie. only contain one signal per cluster, and the signals are hence optimally separated [23].
2.3. Semi-synthetic Database
To measure the performance of the algorithms COMBI, MULTICOMBI, WASOBI and EFICA, compared with more traditional algorithms such as JADE, we built a semi synthetic database [24],[25] as
XAECG and XFECG are 3-D sources representing the maternal and fetal cardiac components [26], Hf and Hm are the volume conduction transfer matrices for the mother and fetus respectively. In this model, the maternal signal HmXMECG is assumed as interference while n is assumed as noises for the fetal signal HfXFECG. The parameters α and β control the signal to interference ratio (SIR) and signal to noise ratio (SNR). To model can be selected from Gaussian white noise or pink noise. The number of rows of Hf and Hm can be adjusted as the number of leads required to simulate the AECG. It is also possible to model the relative fetal position respect to the axes maternal body introducing specific angles between the subspaces of the matrix columns [5]. For the current study, the pink noise have a spectral density function that decreases monotonically with frequency s(f) = 1/f.
2.4. Performance parameters
The parameters α and β in (3) control signal to interference ratio (SIR) and signal to noise ratio (SNR), defined as:
The signal to error ratio (SER) is calculated as measures of fetal signal quality before extraction:
Where XFECG_REAL is the desired signal and XFECG_OBS - XFECG_REAL is the error. Here XFECG_OBS is the estimated source signal and XFECG_OBS and XFECG_REAL should be at the same energy level and phase while calculating the error.
3. EXPERIMENTS AND RESULTS
3.1. Data base
In this paper we used a database built according to (3). XMECG and XFECG were taken from diagnosis database PTB [27] from orthogonal leads Vx, Vy, y Vz. PTB has a sampling rate of 1000 Hz. The signals were pre-processed for baseline wander removal and low pass filters with cutoff frequency 100 Hz. To build a database of 26 records AECG, 52 records from healthy subjects were used, half of which were randomly selected, represent XMECG, and the remaining were re-sampled to 500 Hz to simulate fetal sources XFECG, because the fetal heart rate is typically twice the fetal heart rate. In order to get eight channels of abdominal observations XAECG in (3), were selected random matrix Hf and Hm of 8x3 with angles between the sub-spaces of the column below 40 °.
SIR values were swept in the range of -5dB to -30dB, which are in the range of actual values. SIR = 0 dB, indicates that the FECG has a higher power than the MECG which is not real, therefore is excluded from the analysis. SNR values were swept in the range of 0dB to 30dB. For each noise type and for each algorithm, all possible combinations between the values of SNR and SIR are investigated to 20 repetitions. The step is 5 dB. In each repetitions Hf, Hmand noise are varied randomly. SER is the average obtained for 20 repetitions. At each step of the process the signals are normalized for purposes of calculating the SIR, SNR and SER. All simulations were carried out on data segments of 10 seconds with a sampling frequency of 500 Hz.
3.2. Results
As an illustration, typical signals used for generating an AECG are plotted in Fig. 1-a. The AECG resulting of applying the method according to (3) is plotted in Fig. 1-b. The fetal signal power to maternal interference power ratio SIR is -5dB, and white noise is added so that the fetal signal power to noise power ratio SNR is 20 dB. The result of applying the MULTICOMBI algorithm for separating the MECG and the MECG is plotted in Fig. 1-c.
Each channel of the FECG sources, shown at the top of Fig. 1-a, is compared by the SIR with each channel of Fig. 1-c. Then, the three signals in Fig. 1-c with SIR greater are considered the FECG estimated, as shown in Fig. 1-d.
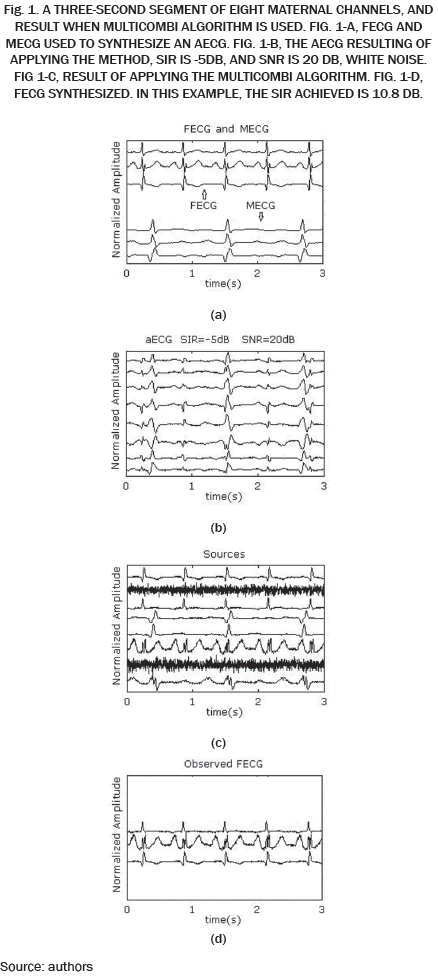
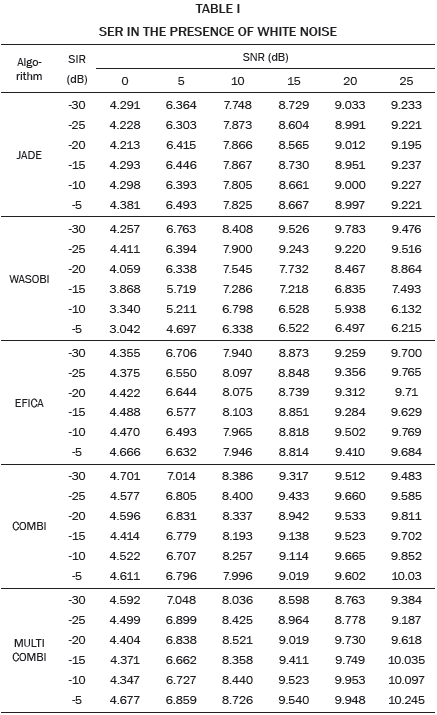
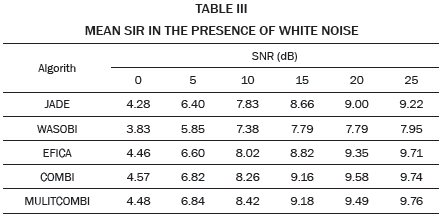
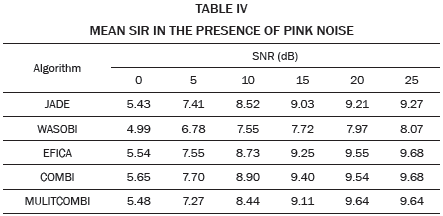
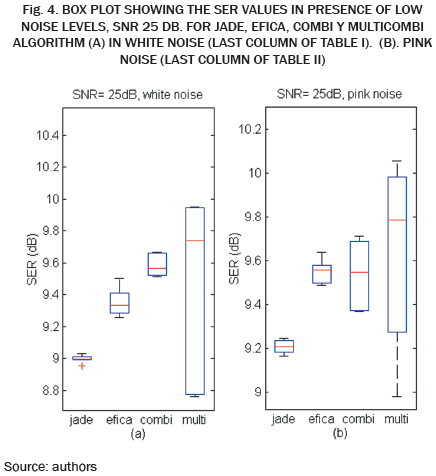
The higher SIR calculated, 10.8 dB in this example, is stored and averaged with the results of the remaining 19 repetitions in which the noise, Hf and Hm, matrix, are changed randomly. The resulting SIR is reported in Table I when white noise is used and in Table II when pink noise is used.
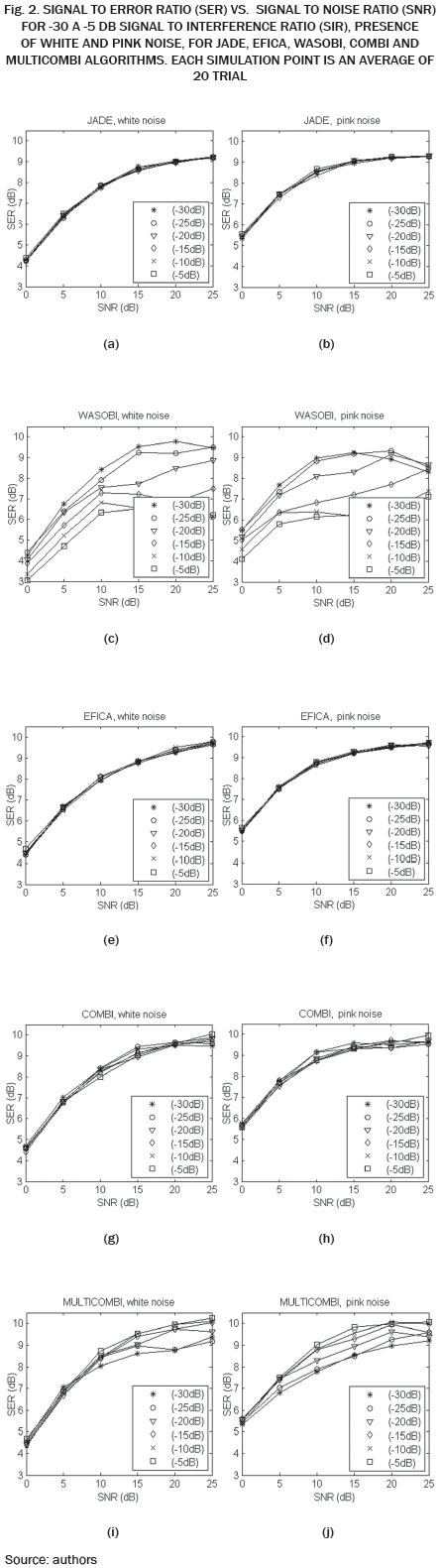
The SER obtained by each algorithm for different combinations of SNR and SIR is shown in Tables I and II. It can be seen in Table I, SER values are slightly lower compared to the values in Table II, indicating that the algorithms are more susceptible to white noise and pink noise least.
The data in Tables I and II are plotted in Fig. 2. Each curve corresponds to a value of SIR in the range -30 to -5 dB. As expected, for high values of noise low SER values are obtained, while for low values of noise high SIR values are obtained. Except WASOBI and MULTICOMBI to a lesser degree, a behavior roughly constant regarding SIR exhibit algorithms analyzed. The mean SER is shown in Tables III and IV.
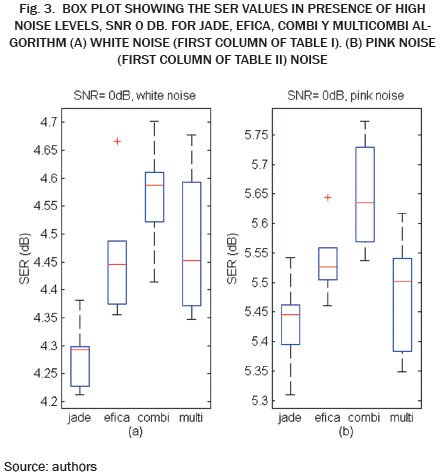
As we can see in Tables I, II, III and IV, and Fig. 2, JADE and EFICA algorithms have high performances while the WASOBI algorithm exhibits the worst performance. This is because the AECG sources are both, statistically independent and not Gaussian with distribution function determined to by its higher order statistics, greater extent, and has a Gaussian distribution function determined by its second-order statistics, lesser extent. Since AECG signals are both Gaussian and non-Gaussian components, COMBI and MULTICOMBI algorithms exploit the strengths of both techniques, exhibiting better performance.
While it is true that the performance of algorithms COMBI and MULTICOMBI are superior, especially in the presence of white noise, it is of standing out that are more dispersed the algorithm MULTICOMBI results. This may be because the clustering scheme used for MULTICOMBI is an ad hoc algorithm. However, for the task of separating the sources in the AECG, situations containing poorly distinguishable clustering can occur. This is, the residual clusters produced by each method separately should overlap, e.g., if there are some similarities in spectra between sources in different clusters of WASOBI or if there are sources which are roughly Gaussian for EFICA [13].
4. DISCUSSIONS AND CONCLUSIONS
In this paper, we have calculated and compared the performance of five algorithms BSS based, in a semi-synthetic database in the problem to extract the FECG. The WASOBI algorithm exhibits the worst performance, but combining both WASOBI and EFICA in COMBI and MULTICOMBI algorithms, the strengths of both techniques are exploited exhibiting better performance. However, are more dispersed the algorithm MULTICOMBI results. This is because MULTICOMBI is an ad hoc algorithm; therefore, the clustering scheme used must be optimized for the task of separating the sources in the AECG.
REFERENCIAS
[1] L. Hui and D. W. Bianchi, "Prenatal pharmacotherapy for fetal anomalies: a 2011 update," Prenat. Diagn., vol. 31, no. 7, pp. 735-743, 2011. [ Links ]
[2] R. A. Shepoval'nikov, A. P. Nemirko, A. N. Kalinichenko, and V. V. Abramchenko, "Investigation of time, amplitude, and frequency parameters of a direct fetal ECG signal during labor and delivery," Pattern Recognit. Image Anal., vol. 16, no. 1, pp. 74-76, 2006. [ Links ]
[3] N. J. Outram, E. C. Ifeachor, P. W. J. Van Eetvelt, and S. H. Curnow, "Techniques for optimal enhancement and feature extraction of fetal electrocardiogram," IEE Proc.-Sci. Meas. Technol., vol. 142, no. 6, pp. 482-489, 1995. [ Links ]
[4] Y. C. Park, K. Y. Lee, D. H. Youn, N. H. Kim, W. K. Kim, and S. H. Park, "On detecting the presence of fetal Rwave using the moving averaged magnitude difference algorithm," IEEE Trans. Biomed. Eng., vol. 39, no. 8, pp. 868-871, 1992. [ Links ]
[5] R. Sameni, M. B. Shamsollahi, C. Jutten, and G. D. Clifford, "A nonlinear Bayesian filtering framework for ECG denoising," Biomed. Eng. IEEE Trans. On, vol. 54, no. 12, pp. 2172-2185, 2007. [ Links ]
[6] W. Zheng, H. Liu, A. He, X. Ning, and J. Cheng, "Singlelead fetal electrocardiogram estimation by means of combining R-peak detection, resampling and comb filter," Med. Eng. Phys., vol. 32, no. 7, pp. 708-719, 2010. [ Links ]
[7] A. Khamene and S. Negahdaripour, "A new method for the extraction of fetal ECG from the composite abdominal signal," Biomed. Eng. IEEE Trans. On, vol. 47, no. 4, pp. 507-516, 2000. [ Links ]
[8] P. P. Kanjilal, S. Palit, and G. Saha, "Fetal ECG extraction from single-channel maternal ECG using singular value decomposition," Biomed. Eng. IEEE Trans. On, vol. 44, no. 1, pp. 51-59, 1997. [ Links ]
[9] V. Zarzoso, A. K. Nandi, and E. Bacharakis, "Maternal and foetal ECG separation using blind source separation methods," Math. Med. Biol., vol. 14, no. 3, pp. 207-225, 1997. [ Links ]
[10] M. Richter, T. Schreiber, and D. T. Kaplan, "Fetal ECG extraction with nonlinear state-space projections," Biomed. Eng. IEEE Trans. On, vol. 45, no. 1, pp. 133-137, 1998. [ Links ]
[11] B. Widrow, J. R. Glover Jr, J. M. McCool, J. Kaunitz, C. S. Williams, R. H. Hearn, J. R. Zeidler, E. Dong Jr, and R. C. Goodlin, "Adaptive noise cancelling: Principles and applications," Proc. IEEE, vol. 63, no. 12, pp. 1692-1716, 1975. [ Links ]
[12] P. Tichavsky, Z. Koldovsky, E. Doron, A. Yeredor, and G. Gómez-Herrero, "Blind signal separation by combining two ICA algorithms: HOS-based EFICA and time structure-based WASOBI." Florence: Eur. Signal Process. Conf. (EUSIPCO), 2006. [ Links ]
[13] P. Tichavsky, Z. Koldovsky, A. Yeredor, G. Gómez-Herrero, and E. Doron, "A hybrid technique for blind separation of non-Gaussian and time-correlated sources using a multicomponent approach," Neural Netw. IEEE Trans. On, vol. 19, no. 3, pp. 421-430, 2008. [ Links ]
[14] J.-F. Cardoso and A. Souloumiac, "Blind beamforming for non-Gaussian signals," in IEE Proceedings F (Radar and Signal Processing), vol. 140, pp. 362-370, 1993. [ Links ]
[15] A. Yeredor, "Blind separation of Gaussian sources via second-order statistics with asymptotically optimal weighting," Signal Process. Lett. IEEE, vol. 7, no. 7, pp. 197-200, 2000. [ Links ]
[16] Z. Koldovsky, P. Tichavsky, and E. Oja, "Efficient variant of algorithm FastICA for independent component analysis attaining the Cramér-Rao lower bound," Neural Netw. IEEE Trans. On, vol. 17, no. 5, pp. 1265-1277, 2006. [ Links ]
[17] A. Belouchrani, K. Abed-Meraim, J.-F. Cardoso, and E. Moulines, "A blind source separation technique using second-order statistics," Signal Process. IEEE Trans. On, vol. 45, no. 2, pp. 434-444, 1997. [ Links ]
[18] I. Hyvarinen, "Fast and robust fixed-point algorithms for independent component analysis," Neural Netw. IEEE Trans. On, vol. 10, no. 3, pp. 626-634, 1999. [ Links ]
[19] D. Taralunga, M. Ungureanu, R. Strungaru, and W. Wolf, "Performance comparison of four ICA algorithms applied for fECG extraction from transabdominal recordings," in Signals, Circuits and Systems (ISSCS), 2011 10th International Symposium on, pp. 1-4, 2011. [ Links ]
[20] K. V. K. Ananthanag and J. S. Sahambi, "Investigation of blind source separation methods for extraction of fetal ECG," in Electrical and Computer Engineering, 2003. IEEE CCECE 2003. Canadian Conference on, vol. 3, pp. 2021-2024, 2003. [ Links ]
[21] R. Romo Vázquez, H. Vélez-Pérez, R. Ranta, V. Louis Dorr, D. Maquin, and L. Maillard, "Blind source separation, wavelet denoising and discriminant analysis for EEG artefacts and noise cancelling," Biomed. Signal Process. Control, vol. 7, no. 4, pp. 389-400, 2012. [ Links ]
[22] M. K. Nath and J. S. Sahambi, "Independent component analysis of functional MRI data," in TENCON 2008-2008 IEEE Region 10 Conference, pp. 1-6, 2008. [ Links ]
[23] P. Waldmann, "Of 'cocktail parties' and exoplanets," Astrophys. J., vol. 747, no. 1, p. 12, 2012. [ Links ]
[24] L. O. Sarmiento Álvarez, A. González, and J. Millet, "Synthetic database for testing algorithms of fetal ECG extraction from abdominal ECG," in Image, Signal Processing, and Artificial Vision (STSIVA), 2012 XVII Symposium of, pp. 56-61, 2012. [ Links ]
[25] R. Sameni, G. D. Clifford, C. Jutten, and M. B. Shamsollahi, "Multichannel ECG and noise modeling: application to maternal and fetal ECG signals," EURASIP J. Adv. Signal Process, 2007. [ Links ]
[26] R. Sameni, C. Jutten, and M. B. Shamsollahi, "A deflation procedure for subspace decomposition," Signal Process. IEEE Trans. On, vol. 58, no. 4, pp. 2363-2374, 2010. [ Links ]
[27] A. L. Goldberger, L. A. Amaral, L. Glass, J. M. Hausdorff, P. C. Ivanov, R. G. Mark, J. E. Mietus, G. B. Moody, C.-K. Peng, and H. E. Stanley, "Physiobank, physiotoolkit, and physionet components of a new research resource for complex physiologic signals," Circulation, vol. 101, no. 23, pp. e215-e220, 2000. [ Links ]