1. INTRODUCTION
Contribution of agriculture towards the economy of any developed and developing nation is significant. Agriculture serves as the most important and peaceful source of livelihood and plays a major role in generating national income through its productivity. It also provides employment for the people in rural areas and offers food security to the nation. Agriculture is directly or indirectly contributing to food supply, clothing, medicine and adds revenue through international trade as well. Hence the increase in productivity enhances the growth of a nation.
There is an increased demand for food nowadays due to the rapid increase in population which has led to the need to increase the crop yield which remains an open research challenge that needs to be addressed irrespective of the crop type. There are several internal and external factors that affect the yield of the crops. Some of the internal or genetic factors that impact yield are quality of grains, their chemical composition, tolerance towards water salinity, insects, diseases, etc. The environmental or external factors include the climatic conditions such as temperature, humidity in atmosphere, velocity of wind, presence or absence of vital atmospheric gases, soil fertility, drought [1], etc.
Diseases in plants are majorly due to viral, fungal and bacterial infections along with insects. Early detection of plant disease is the need of the day to increase the productivity of the plants. Due to the advances made in the technology, computer vision and image processing techniques are used nowadays in detection of plant diseases. Among all the machine learning techniques, deep learning has evolved as a formidable technique that contributed significantly in all most all domains especially in image related problems in agriculture as well [2]. With its deeper network and having a stack of convolutional layers, deep learning architectures can learn features from images in a hierarchical fashion. Applications of deep learning in agriculture are widely spread: leaf-plant disease detection, land-cover, crop type classification, crop and its yield identification and estimation, fruit counting, identification of weeds, seeds, pest detection are some of them.
Extracting essential features to discriminate diseases with various symptoms and spots is required for any disease classification task. Traditional hand-engineered feature selection is time-consuming and performance will rely on their selection as well. Deep learning is the optimal solution for this problem, which automatically extracts and learns the features that enable the classifier to discriminate the images into the classes they belong to. It is very much significant to protect and prevent the plants from diseases and early detection is as important as other factors involved. This motivated us to develop an automated disease-detection system for plants in order to increase their yield.
The contribution of this study is as follows:
We proposed a plant disease identification network, PDDNet-cv, that classifies diseases affecting the plants.
We have conducted an extensive set of experiments to research the best suitable network using transfer learning.
We have carried out a detailed analysis of the performance of all the networks applied for disease detection in crops.
The paper is organized as follows: Section 2 shows the detailed background and survey of existing works in disease detection using deep learning. Section 3 presents the proposed methodology, wherein Section 4 introduces the implementation specific details of the proposed work and the results obtained from various architectures experimented. Section 5 finally concludes the paper with a brief summary and future scope.
2. LITERATURE SURVEY
Detecting diseases in plants through a traditional approach, say manually, has been side-stepped by automatic approaches nowadays. Without the prior knowledge of the diseases in a particular domain of plants, disease detection turns into a difficult task. Hence, we need automatic methods to identify diseases in order to handle and treat them by applying appropriate pesticides. In general, disease-detection techniques fall into two categories: direct and indirect methods. Direct methods include serological and molecular methods [3], whereas the bio-marker and via plant properties are categorized as in-direct methods. Our proposed PDDNet-cv approach can be classified as a direct method of disease detection using imaging techniques. Linear regression models, principal component analysis, discriminant analysis, self-organising map neural network, statistical classifiers and artificial neural networks are some of the widely-used disease-detection methods so far.
Plants are vulnerable to several diseases such as those that affect the roots of the plant and those that affect zones above the roots, namely stems and foliage. Among the diseases that generally affect plant leaves are early blight, late blight, leaf mold, bacterial spot, target spot, septoria leaf spot, two spotted spider mite, mosaic virus and yellow leaf curl virus. In [4], citrus leaf lesions are detected and classified through color, texture and geometric features. These features are fed to Support Vector Machine (SVM), as a multi-class classifier. Leaf lesions are initially segmented through weighted segmentation techniques using chi-square and threshold methods; from the segmented lesions, the features are extracted for classification. The texture features include Haralick features, cluster prominence and shade, homogeneity, and energy whereas the lesion shape, size, and position area are few of the geometric features used for classification. The challenge here with the work done in [4] is the manually engineered methods that are used to extract the necessary features. Moreover, the manual feature engineering will be time consuming and also will need more domain knowledge and expertise.
In order to automate the classification of diseases in plants, several machine learning approaches have been widely used. With the emergence of high configuration systems, deep learning approaches have found their light again. Deep learning has contributed in an extensive way in recognizing very large-scale images and has also found its applications in agriculture. Plant disease detection and plants classification are two significant applications where deep learning algorithms are widely used to automate the processes in agriculture. Convolutional Neural Networks (CNN) are the popular choice for image classification in deep learning. CNN helps extract the features automatically and overcome the difficulties in choosing the appropriate features from images in manual feature engineering. A CNN architecture with five convolutional layers and two fully connected layers are used in [5] to determine the severity of the plant disease in apple black rot images. The authors have also used transfer learning on VGGNet, Inception-v3, and ResNet50 by freezing all the layers of the three pre-trained models to detect the four stages of severity, namely healthy, early, middle and end stages of diseases.
[6] is another work where 13 different types of plant diseases are classified using deep learning architecture. The authors used images of different disease-affected plants downloaded from internet and augmented to increase the number of images for training their model. The accuracy obtained was 96.3%. For evaluating, the methodology benchmark dataset is not used. Also [7], [8], [9], [10] are some of the other works that have used CNN for disease classification task. Transfer learning is one another domain in machine learning, where one can reapply and reuse the knowledge gathered earlier, for solving problems in similar domains. Instead of training the network from the beginning, transfer learning allows the weights of the already trained model to be transferred to the new model [11] and also offers provisions for fine tuning them.
Deep learning requires a large amount of input samples to get the training done, whenever the training samples are not sufficient, learning acquired by pre-trained models on similar tasks can be utilized. Rebuilding a new model will take too much time, space and computational power [12], whereas the model built through transfer learning can help enhance the performance of the model as well. When we fine-tune the pre-trained model, one can freeze all the layers of the pre-trained model and our data can be tested on it or we can freeze the layers partially and train the rest of the layers with our training data. There are several pre-trained models available, some of them are VGG [13], ResNet [14], GoogLeNet [15], different versions of Inception, Inception-ResnetV2 [16], AlexNet [17], etc.
A pre-trained InceptionV3 model has been used in [18] for three classes of disease detection in Cassava plants. Cassava mosaic disease, red mite damage, brown leaf spot are the diseases that have been detected by their proposed classifier. The results obtained through this pre-trained classifier have been compared with KNN and SVM classifiers. GoogLeNet was used for disease identification from individual lesions and spots for around 14 plants such as coconut, cashew, citrus, cassava, corn, cotton, coffee, etc. The limitation of deep learning networks for classifications is the need of large dataset for training -as observed in [19]and hence image augmentation techniques were applied to increase the images of each class. 57 diseases of 28 plants are classified using pre-trained models VGG, AlexNet, and GoogLeNet in [10]. In [20], the authors have investigated the impact of size and variety of images on deep learning architectures for classification of diseases on plants. GoogLeNet, a pre-trained model, has been used, where the effect of removing the small background, positive and negative backgrounds was studied. Mixed responses have been observed among different plant varieties with respect to the removal of backgrounds.
3. PROPOSED METHODOLOGY
This research focusses on developing a generic framework for detecting and identifying the presencia of disease in the crops.
In particular,
A deep-learning-based system to accurately detect the diseases in crops such as Tomato, Potato and Bell Pepper.
To apply transfer learning concepts for disease detection
3.1 System Architecture
Due to the characteristics of image, identifying a suitable feature to discriminate various diseases plays a major role. The human perceptual way of recognizing the plant diseases from leaf images is based on pixel intensity distribution, texture properties and few other cues. In early days, appearance of the leaves helped the farmers to identify the type of disease and the pesticide to be used, because of having abundant knowledge and expertise. The future of agriculture is moving towards digitalized smart agriculture. In order to automate the process of visual feature extraction and identifying the category of diseases, a deep-learning-based network is proposed.
The challenge is that the images of the disease-affected plant leaves across the classes look similar, but differ within the same class, so that human eyes fail to detect the different types. In conventional machine learning, one needs to choose and extract the suitable and appropriate features to discriminate the diseases of plants, which will be too complex on these types of images. In order to handle this, we are in need of an automatic feature extracting system, which extracts the necessary features to discriminate the images.
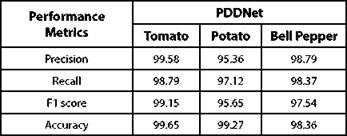
In this research, we designed a deep learning architecture named PDDNet (Plant Disease Detection Network) to identify / detect the category of diseases in plants. Evaluating the model with one test set will not be reliable, since the accuracy of the model differs when it is tested with different test sets due to sample variability among the training and test sets. Also, in order to capture the underlying patterns and to train the model with all the samples in the dataset so we can build a generalized model, cross validation is used in PDDNet and hereafter will be referred to as PDDNet-cv (Plant Disease Detection Network with cross validation).
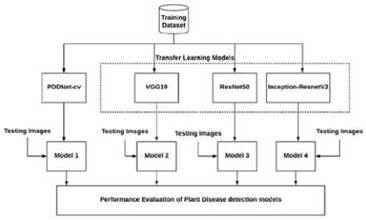
Deep learning networks require huge lot of labelled data for training, however, getting labelled data is a tedious process. This led us to apply transfer learning, where we use the pre-trained models for classification. The pre-trained models used here are experimented with different parameter settings to study their performance. Then, performance of PDDNet-cv is evaluated against the pre-trained models. The overview of the plant disease detection architecture is shown in Fig. 1.
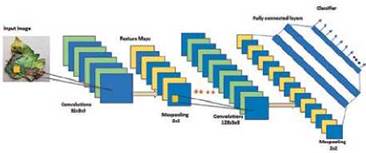
Our proposed discriminative network, PDDNet-cv has a sequence of convolutional layers with activation functions, pooling layers followed by dense layers as shown in Fig. 2. PDDNet-cv has six convolutional layers with 32, 64, 128, 128, 512 and 1024 filters respectively. Each convolutional layer produces a feature map by moving the filter/kernel/ weights over the image which serve as the receptive area. These kernels will be automatically initialized and get updated during the backward pass of the network based on optimizing function. Relu is used as the activation function across all the convolutional layers since Relu has proven to overcome the vanishing gradient problem exhibited by other conventional activation functions in literature.
The pooling layer that follows each convolutional layer serves to downsize the number of parameters and hence the corresponding computation incurred will also get reduced. The pooling layer also helps in controlling the over-fitting problem. The fully connected layers carry out the classification of diseases from the feature maps generated by the convolutional layers. The first and second dense layers have 1024 outputs each with dropouts of 50% and 20% following the dense layers. The dropouts are one of the other factors that help the network from over-fitting. The last dense layer has n classes corresponding to the number of classes. The architecture of PDDNet-cv is shown in Fig. 2.
The feature maps computed from the sixth layer are received by the first dense layer with 1024 nodes. The weight vectors were learned from the dense layers for prediction. The last dense layer is designed with the appropriate nodes for the diseases. By backpropagation, the weight vectors get updated and are learned for accurate detection of diseases.
3.2 Transfer Learning
Transfer learning refers to the process of reusing a model learnt for a given task for similar tasks. Our second objective in PDD is to identify the best performing pre-trained transfer learning model. Due to the unavailability of labelled data for solving problems, transfer learning plays a role. Transfer learning is popular in deep learning literature, where pre-trained models such as VGG16, VGG19 [13], AlexNet, GoogLeNet. SqueezeNet, ResNet50 [14] and Inception-ResNetV2 [16] have been developed and are reused in computer vision and NLP tasks.
For computer vision tasks, many pretrained models have been developed so far, among them VGG19, ResNet50 and Inception-ResNetV2 are the models explored for predicting disease in this work. VGG19 is designed by Visual Geometry Group (VGG) of University of Oxford which has secured second place in ImageNet classification challenge in 2014. It has 19 layers arranged in 5 blocks where each block has stack of 2, 2, 4, 4 and 4 convolutional layers. Image size accepted by VGG19 is 224 x 224, the receptive fields or the kernel size of each of the convolutional layer is 3 x 3, the first 2 fully connected layers have 4096 nodes where the last one has 1000 nodes, as it was designed for ImageNet Large Scale Visual Recognition Challenge (ILSVRC).
The second pre-trained transfer learning model used for disease detection is residual network which facilitates residual learning through skip/shortcut connections. When the deep neural network goes deeper, the accuracy somewhere gets saturated and also sometime gets deviated. This is solved and eliminated using ResNets. ResNet50, as the name suggests, has 50 layers with 49 convolutional layers and 1 fully connected layer. It has a stack of non-linear convolutional layers (Y=F(x)) and an identity mapping function where the input is mapped as output. A ResNet block in this architecture has 3 layers in depth, where the skip connection is done before a Relu layer which is found to have enhanced results.
The third pre-trained model chosen for PDD is Inception-ResNetV2, which is a hybrid of Inception module of GoogLeNet and the residual blocks of ResNet architectures. It has 3 Inception-ResNet blocks with a total 164 layers in it. Each interior inception grid has 35 x 35, 17 x 17 and 8 x 8 grid modules with reduction blocks that modify the height and width of the grids.
All the three pre-trained transfer learning models are used in three different ways for disease detection, namely:
If the feature maps for the source task and the target task were similar, the same pre-trained architecture could be used.
Feature maps may not be similar for source and target task and if the target task has sufficient labelled data for training, simply adapt the architecture of the pre-trained model and learn the feature map using the target task data.
Third category is to tune the pre-trained model architecture as per the target task by freezing few layers of the architecture.
In this research all the three forms of transfer learning approaches were explored for plant disease detection as target task. Analysis of all the pre-trained approaches is discussed in detail in the next section. In all the transfer learning models, a final fully-connected layer with the intended number of disease classes has been added in order to customize them for PDD task.
4. EXPERIMENTS AND RESULTS ANALYSIS
4.1 Dataset description
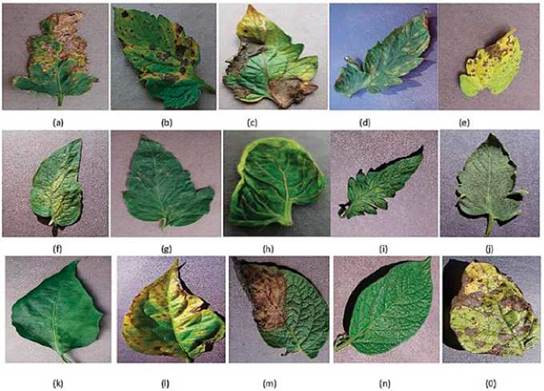
PlantVillage dataset [21] is used for disease detection on plants from which Bell Pepper, Potato and Tomato have been used in this work. There are a total 7166 tomato leaf images across 10 classes for Tomato, 2575 images of Bell Pepper leaves and 2153 images of Potato plants present in PlantVillage dataset. The labels of the classes for Tomato leaves are Bacterial Spot, Early Blight, Late Blight, Leaf Mold, Septoria leaf Spot, Two spotted Spider Mites, Target Spot, Yellow Leaf Curl Virus, Mosaic Virus and Healthy. Early Blight, Late Blight and Healthy are labels corresponding to Potato leaves and Bacterial Spot, Healthy are the class labels of Bell Pepper leaves. Sample leaves of all classes are shown in Fig. 3. A proposed system for disease detection is implemented in Python3 using the deep learning library Keras. Nvidia GPU GTX1080 Ti with 11 GB graphics card in Linux OS: Ubuntu 16.04 LTS and also the Google colab were used for implementation and to carry out the different experimental runs.
Fuente: Authors.
Fig. 3 TOMATO LEAVES AFFECTED WITH DIFFERENT DISEASES (A) BACTERIAL SPOT, (B) EARLY BLIGHT, (C) LATE BLIGHT, (D) LEAFMOLD, (E) SEPTORIA LEAF SPOT, (F) TWO SPOTTED SPIDER MITE, (G) TARGET SPOT, (H) MOSAIC VIRUS, (I) YELLOW LEAF CURL VIRUS AND (J) HEALTHY (K) BELL PEPPER HEALTHY, (L) BELL PEPPER BACTERIAL SPOT, (M) POTATO EARLY BLIGHT, (N) POTATO LATE BLIGHT AND (0) LATE BLIGHT HEALTHY
4.2 Experiments conducted
Various experiments conducted are listed below:
Application of PDDNet on the chosen dataset for performance evaluation
Application of PDDNet-cv
Evaluating Pre-trained transfer learning models (VGG19, ResNet50 and Inception-ResNetV2,) with following conditions:
Training all the layers of pre-trained models with PlantVillage dataset.
Partially training transfer learning model with chosen dataset by freezing few layers of them.
Using the transfer learning model without training the network with chosen target dataset.
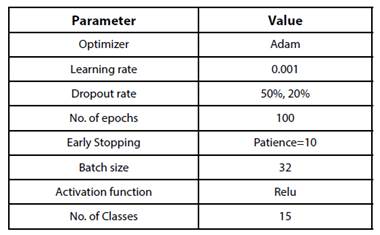
All the models were evaluated for 100 epochs with a batch size of 32.
4.3 Experiment 1: PDDNet
The proposed PDDNet model is experimented with the PlantVillage dataset for disease prediction. This network involves 6 convolutional and 3 dense layers. To avoid the problem of overfitting of the proposed convolutional network, Dropouts and Batch Normalization have been used. Batch normalization helps each layer to learn independently of previous and other layers. It also helps in accelerating the learning process with high learning rate during the training. The fine tuning of the architecture was performed by varying the hyper-parameters. Image augmentation has been applied by performing transformations such as rotation, shifting, shearing and horizontal flip.
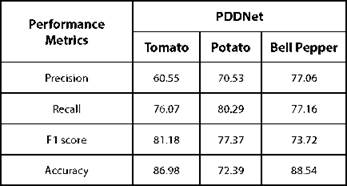
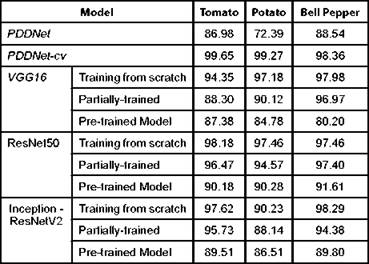
As one of the best practices, checkpointing is employed which enables the model parameters to be saved every epoch. Early stopping is also implemented to avoid over-fitting the training data. The hyper-parameters to be learned during training are listed in Table I. Validation set is used to tune the hyper-parameters, so that over-fitting could be reduced. The model thus constructed is evaluated with the test set. When PDDNet is evaluated with Plant village dataset, it has obtained an accuracy of 86.982% -other metrics are tabulated in Table II. This simple deep learning model PDDNet when applied on plant village dataset has reported poor accuracy in detecting diseases in Bell Pepper plant as compared to other two crops. The accuracy obtained is comparatively very poor for this model, when compared with the other models built. The reason is very clear: the model built is not generalized because of the presence of unseen data. This problem of overfitting is handled by using cross validation and is implemented as PDDNet-cv.
4.4 Experiment 2: Simple PDDNet vs PDDNet-cv
In order to enhance the performance of detecting diseases, k-fold cross validation is introduced in our model and referred to as PDDNet-cv. To estimate and to improve the model performance, the basic architecture undergoes k-fold cross validation which can handle the challenges of unseen data. Due to this, the error is averaged across all the k trials that can enhance the efficiency of the model which can also handle the issue of overfitting. The accuracy of detecting diseases has been improved significantly using PDDNet-cv. Detecting diseases associated with tomato plant has been improved with an accuracy of 99.65%. The proposed PDDNet-cv has given a better performance than the initial model PDDNet, which is without cross validation. It is learnt that the cross validation has improved the performance among all the crops extensively which has allowed the model to learn all the discriminating features well enough to identify the diseases properly. The margin of difference in classification accuracy of PDDNet-cv among the three crops is minimal. Though the leaves of the crops in Plant village dataset appeared to be very similar, our proposed PDDNet-cv has performed well when compared with the architecture without cross validation PDDNet. The lack of performance in Bell Pepper was due to the presence of a smaller number of samples in the dataset. The performance measures of PDDNet-cv were reported in Table III and in Table IV.
4.5 Experiment 3: Transfer Learning models
All three pre-trained models, namely VGG19, ResNet50 and Inception-ResNetV2 networks, are adapted as specified in section 3.2, training all layers from scratch, partially freezing few layers and freezing all layers as a pretrained model.
4.5.1 Training from scratch
Various deep learning models, pre-trained with ImageNet such as VGG19, Inception-ResNetV2 and ResNet50, are used in this study for disease's detection. Instead of using the pre-trained weights of these architectures trained on ImageNet dataset, the Plant village dataset is used for training. The classifier layer of these models is removed and new fully connected layers are added with required number of classes for disease classification. Among these models, ResNet50 performed well with an average accuracy of 97.68%. The results of training from scratch using the three base models are reported in Table IV. This leads to the observation that the global pooling in ResNet50 might have enhanced the classification accuracy, though the depth of ResNet50 model (168 layers) when compared with Inception-ResNetV2 (572 layers) is inferior. Among the other two pre-trained models, VGG19 has come closer to ResNet50 with 96.5% of accuracy in average on the Plant Village dataset.
4.5.2 Partially-trained
VGG19, Inception-ResNetV2 and ResNet50 are partially trained by freezing first few sets of selected convolutional layers in each of the architectures. Choosing the number of layers to freeze was done through varied set of experiments. The results are tabulated here in the Table IV. Tuneable parameters of these three classifiers are varied and an empirical study was done to identify the best model. Performance of these models are found to be good when we freeze the first 10 layers in VGG19, the first 70 layers in ResNet50 and the first 100 in Inception-ResNetV2. The results show that ResNet50 leads in performance with an average accuracy of 96.15% than the other two base models.
4.5.3 Transfer learning via pre-trained models
This approach has used all three pre-trained models as inference models where the weights of the models trained on ImageNet dataset have been used as such and kept constant during target dataset classification. The final classification layer in VGG19, Inception-ResNetV2 and ResNet50 is replaced with new fully connected layers with nodes corresponding to the diseases in target dataset. In this approach the Plant Village dataset has been used for testing the models as the target dataset. This approach has achieved 90.69% of average accuracy by ResNet50. The misclassification errors are due to the difference in type of data between the target and source dataset, which are not similar in this study.
4.5.4 Observations on using transfer learning models
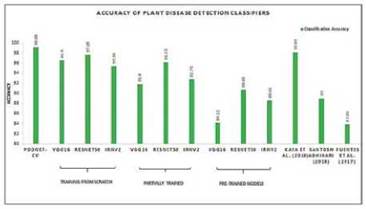
The average accuracy of the transfer learning models on Plant Village dataset is shown in Fig. 4. The inferences that can be observed from the results of transfer learning approaches are as follows:
Fuente: Authors.
Fig. 4 AVERAGE CLASSIFICATION ACCURACY OBTAINED ACROSS ALL CLASSIFIERS ON PLANT VILLAGE DATASET
The ResNet50 had the best performance in classifying the diseases of Plant Village dataset followed by the Inception-ResNetV2. The reason behind their best performance will be because the skip or shortcut residual connections present in these two models enabled them to perform better than the VGG19 pre-trained model. Even though the depth in layers in VGG19 is less than ResNet50 and Inception-ResNetV2, skip connections in ResNet50 has enabled the network to learn global features.
Training the base models from scratch obtained the best performance out of the three approaches of transfer learning.
Using the pre-trained models as inference system is not suitable for this study. The reason being that the data similarity between source and target datasets is very less.
Partially-trained transfer learning is observed to be a challenging approach in our study, since selecting the layers for learning is a difficult task and it is not possible to identify the optimum layers.
In order to show the statistical significance in the classifying ability of the three pre-trained classifiers, t-test was conducted mutually between them. The p values show that there is a significant difference among the three classifiers with t stat and t critical are {1.7173, 2.1788}, {0.5696, 2.1447} and {1.4313, 2.1314} respectively.
4.6 PDDNet-cv vs best transfer learning approaches
PDDNet-cv has outperformed all the variants of the transfer learning approaches considered for this study, in its performance in individual crops and as well as in average. Our proposed architecture has obtained 99.09% as average accuracy, where ResNet50 has achieved 96.52%, when it was trained from scratch and the same is shown in Fig. 4 and in Table IV. It is observed that shallow network is sufficient enough for disease detection task. The transfer learning models have high misclassification errors due to the overfitting that has been avoided in PDDNet-cv with a smaller number of convolutional layers to represent features and the dense layers to classify them.
4.7 Comparison of PDDNet-cv with existing methods
Performance of our method PDDNet-cv with 99.09% average accuracy, is compared against the state-of-the-art methods [9, 12, 22]. The average accuracy of detecting diseases in plants, is shown in Fig. 4. When compared to [12], our methodology has outperformed it in discriminating diseases. Two networks are proposed in [12], the first one being CNN-RNN which has VGG16, AlexNet as feature extractors followed by an RNN and the second architecture is an end-to-end CNN with 15 convolutional layers. Such deep layered architectures are not needed for this task of disease detection; this has been observed through the performance of PDDNet-cv.
With regards to [9, 22], PDDNet-cv has significantly performed well. A classifier similar to YOLO was used in [22], which has 24 convolution layers and 2 fully connected layers and has achieved 89% of accuracy, in [9] three detector architectures were used, namely Faster Region-based Convolutional Neural Network (Faster R-CNN), Region-based Fully Convolutional Network (R-FCN), and Single Shot Multibox Detector (SSD). Faster R-CNN with VGG16 has gotten 83.06%, R-FCN with ResNet50 has obtained 85.98%, whereas SSD with ResNet50 has achieved 82.53%. These comparisons have shown that, with the kind of images in PlantVillage dataset, the classification network need not have too many layers stacked up, a shallow network is sufficient enough to discriminate the diseases. Our proposed, PDDNet-cv has achieved significant performance with few feature extraction layers and dense layers.
5. CONCLUSION
In this paper we have designed a specialized deep learning network PDDNet-cv for plant disease detection. The classifier we have proposed has detected 15 different classes of diseases automatically and has the ability to classify them with an average of 99.09% accuracy on the leaf images of PlantVillage benchmark dataset. The advantage of using PDDNet-cv is that the features are extracted automatically rather hand-engineered. We hope that, through this work, we have made a noticeable contribution in the field of disease detection in plants that may help the individuals who wish to do farming and with good yield, but lack farming expertise. We have also experimented with 9 different classifiers of pre-trained models and their results are observed for disease detection. All the models are empirically tried by varying several tunable model parameters and the best performing fine-tuned models are reported in this study. Better results are observed only when the entire model is trained again with the target dataset.
PDDNet-cv can be further trained with other crop images and can also be used to detect the diseases in those plants. This requires the PDDNet-cv to be fed with few more thousands of additional images and increasing the number of classes in the final layer of the model. As an extension, the number of images could have been increased further, the model parameters could be tuned empirically in order to enhance the classifier performance. This system will have a high impact on sustainable agriculture by increasing the yield of any crop, through early prediction of the possible diseases and treating them on time.