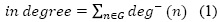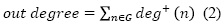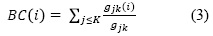Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Nova
versión impresa ISSN 1794-2470
Nova vol.14 no.25 Bogotá ene./jun. 2016
Metabolomics and pesticides: systematic literature review using graph theory for analysis of references
Metabolómica y Pesticidas: Revisión sistemática de literatura usando teoría de grafos para el análisis de referencias
Martha Zuluaga1, Sebastian Robledo2, German A. Osorio-Zuluaga2, Laura Yathe1, Diana Gonzalez1, Gonzalo Taborda1.
1 Universidad de Caldas (Colombia), Facultad de Ciencias Exactas y Naturales
2 Universidad Nacional de Colombia, Facultad de Administración, Bogotá, Colombia
Corresponding autor: martha.zuluaga@ucaldas.edu.co
Martha Zuluaga: 0000-0003-1720-8476, Sebastian Robledo: 0000-0003-4357-4402, Germán A. Osorio: 0000-0002-5613-5858, Gonzalo Taborda: 0000-0003-4358-1506, Laura Yathe: 0000-0002-24'8-8672, Diana Gonzalez: 0000-0002-67'5-8457
Recibido: 17-12-2016 Aceptado: 05-05-2017
Abstract
The systematic literature review presented here is designed to illustrate the role of metabonomics and metabolomics in pesticide exposure studies. The search was conducted in Thomson Reuters Web of Science (ISI Web of Knowledge) database. The references and citations for each article were downloaded for analysis. Graph theory was used to determine relevant articles and distinct relationships between classic and current research in this field through its structural characteristics. The initial network included 4423 nodes and 4978 links, from which indegree, outdegree and betweenness indicators were extracted. After preprocessing the data, the network was reduced to 415 nodes and 974 links. From this network, 80 articles with the highest score between the three indicators were extracted for review. This methodology allowed for the identification of different perspectives of metabolomic and metabonomic pesticide studies that included the mode and mechanism of action, toxicological and biological monitoring, environmental metab-olomics, metabolism, dose response and biomarkers and its role in pesticide exposure.
Keywords: Metabonomics; metabolomics; pesticides.
Resumen
La revisión sistemática de literatura presentada a continuación tiene como objetivo dar a conocer el rol que han tenido la metabolómica en el estudio de la exposición a plaguicidas. La búsqueda se llevó a cabo en la base de datos Thomson Reuters Web of Science (ISI Web of Knowledge). Posteriormente, se descargaron todos los registros producto de resultado de la búsqueda y cada citación dentro de cada artículo. Estas referencias fueron analizadas mediante la teoría de grafos con el fin de identificar los artículos más relevantes, los artículos clásicos y recientes y los que presentan mayor intermediación en el tema de investigación. La red de citaciones fue construida con inicialmente con 4423 nodos (artículos) y 4978 enlaces (citaciones) a los cuales se les determinó los indicadores de grado de entrada, grado de salida y centralidad en el grafo. Posteriormente, esta red de citaciones fue procesada eliminando los artículos desconectados, reduciendo la red a 415 nodos y 974 enlaces. De esta red ya procesada se extrajeron 80 artículos que presentaron mayores indicadores de grado y centralidad. Finalmente, esta metodología permitió la identificación de diferentes perspectivas de los estudios metabolómicos y metabonómicos en la exposición a plaguicidas que incluyen estudios de modo de acción, mecanismos de acción, monitoreo biológico y toxicológico, metabolómica ambiental, metabolismo, dosis respuesta e identificación de biomarcadores.
Palabras clave: Metabolómica, metabonómica, pesticidas.
Introduction
Numerous studies in the fields of industry, environment and health science are related to pesticides (1, 5). Despite the fact that many of the persistent organic compounds have been removed from use or are regulated in the global market (Basel Convention 1992; Rotterdam Convention 2004; Stockholm Convention 2001), their use worldwide is increasing gradually as human population and food requirements increase (6) and for control of infectious vectors. Therefore, bioaccumulation and biomagnification remain a source of health alert.
Nowadays, monitoring pesticide exposure is mainly followed through chromatography because of the development of increasingly more sensitive and robust analytical techniques and methods. However, monitoring pesticide residues is still a challenge as they are present in a very low concentration sometimes already transformed or conjugated with other molecules, making them difficult to separate and quantify. Moreover, the wide spectrum of mixtures and the hundreds of formulation presented in the market, make the analysis of the exposure too complex (6).
In this vein, identification of new biomarkers, pathways analysis, modes and mechanism of action is an alternative to evaluate the exposure event. For this, a growing trend is the bottom up studies like between metabolites in a biological system that helps to explain the momentum of a system metabolomics and metabonomics, because it is focused to identify, quantify and evaluate the interactions, recreating the map of the biochemical interaction in a molecular level (7, 9). For this reason, our principal goal with this review is to report the main findings in metabolomics and pesticide exposure. As well as identify the chemical procedures, software for data processing and data analysis strategies mostly used and the integration with the pathway and network analysis and the role of metabolomics in pesticide exposure.
For this, the review was followed as a systematic literature review and then integrating the results of the search and each citation in a network, in which graph theory analysis was applied. It utilizes three indicators extracted from the graph, indegree, outdegree and betweenness, in order to identify classic and current articles that are more connected within the network and the articles that connect the classical with the recent articles by the shortest path respectively, clusters or communities in this field, among other indicators of the graph.
Search equation and articles selection through network analysis
The search was conducted in the Thomson Reuters Web of Science (ISI Web of Knowledge) database for the time period from January 2001 to June 17, 2015. The terms included in the search equation was used in response to the question: What has been the contribution of metabolomics and meta-bonomics studies to pesticide exposure?
- Topic=(pesticide*) AND Topic= (metabolomic* OR metabonomic*) Refined by: Document Type= (ARTICLE OR REVIEW) Timespan: All years. Indexes: SCI-EXPANDED, SSCI, A&HCI.
The references provided by the search were extracted with all citations, which were then integrated into a network. This procedure can incorporate articles from various sources, regardless of the source database or publishing journal. The methodology proposed by Robledo-Giraldo et al. (10, 11), was used to identify relevant articles.
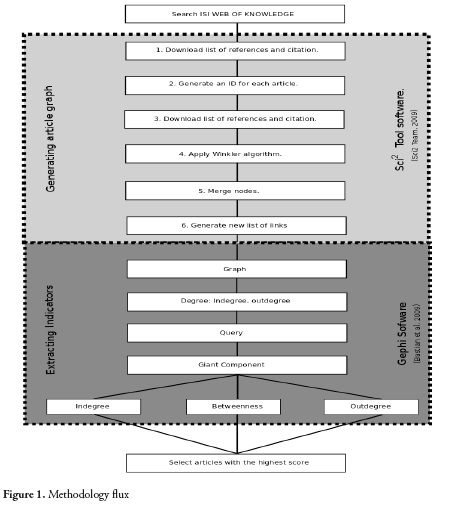
Graph theory was used in order to generate a map that show the articles and the interaction between them and extract relevant information from the topology and characteristics of the network. For this, two open-source software were used: Sci2 tool (Sci2 Team. 2009) (https://sci2.cns.iu.edu) where the list of references and citations were transformed in a network and, for visualization and statistics Gephi (12) (https://gephi.github. io). The methodology flux is shown in figure 1.
Once the network was generated in Sci2 tool, the file was visualized in Gephi and all the networks indicators were calculated on it (figure 1). In-degree is the number of papers that have cited a particular article (Equation 1). Out-dedree is related to the links leading away from the node (13) in this case, all the citations mede in an article but present in the graph (Equation 2).
Wheren, represents the nodes of the graph and G -the graph.
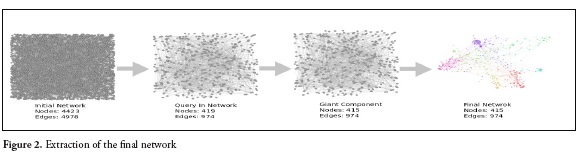
Then, the giant component is calculated. It is defined as a group of nodes that are all connected to each other, directly or indirectly (14). It is a filter used to remove the communities or nodes that are not connected with the main community (Figure 2).
Betweenness Centrality, tallies the number of shortest paths in a graph that pass through a given node; that node has high centrality because it is between many pairs of nodes (Equation 3) (15). Betweenness Centrality, as intuitively indicated by the term, focuses on how close a node is to all the other nodes in a network (16). High values of betweenness represent the articles that connect in a shorter path the classical with current articles.
Where g is the shortest path from node j to k. Thehgorithmdeveloped by Blondel et al. (2008) (17) was applied to the final network to identify clusters of metabolomics and metabonomics studies on pesticides. Modularity class algorithm allows to visualize the partition of a network into communities of densely connected nodes, with the nodes belonging to different communities being only sparsely connected (17).
Network and metrics
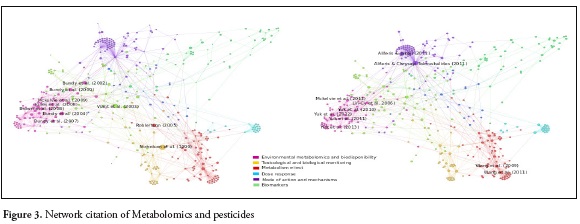
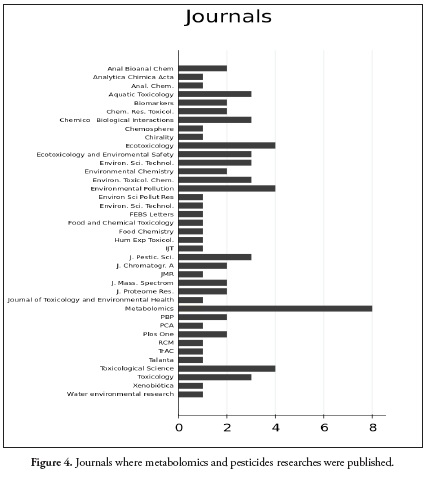
The results of the search equation yielded a network of 4423 nodes and 4978 links. After preprocessing the data (Figure 1.), the network was reduced to 415 nodes and 974 links (Figure 2.) From this final network, the highest score of each indicator (in-degree, betweenness and out-degree) were selected. 80 articles in total were chosen for the purpose of this review. Twenty articles with the highest in-degree, twenty with the highest Betweenness. Thirty with the highest out-degree and ten reviews were found. Those articles represent relevant information from a very wide period of time, extracted from the network. Articles with greater indegree values represent classics and those with higher outdegree values represent the most current articles that are most connected within the network. High betweenness score, represents the articles that made a connection between the classical and the current articles. Figure 3. Show the network and the size of the nodes represents the degree of citation. Figure 3a, the larger nodes are classic articles that have been cited most frequently. Figure 3b shows the most current articles that have been cited to a greater degree in articles within the network and figure 4 shows the journals in which metabolomics and pesticide exposure papers have published.
In the graph visualization, it was possible to identify six different communities or clusters based on different perspectives or approaches of metabolomics studies applied to pesticides (Figure 3). Modes and mechanisms of action 19.3%, biological and toxicological monitoring 17.6%, environmental metabolomics and bioavailability 16.1%, metabolism 15.9%, biomarkers 8.9%, dose response 5.5%. Each perspective will be discussed further.
Classical articles
As described above, articles with the highest in degree indicate those that have been more cited and are consider the classics. Although these classic articles could be considered very old, they present a valuable information and we will make a brief tour through them and describe the main works and findings made this field grow through time. Pioneering work Metabolomics focused on establish robust protocols for sample preparation, to present the chemometric data processing and propose alternatives for future bioinformatic developments for treating either signals NMR or MS, in order to establish the how and the scope of studies.
In metabolomics and metabonomics field the oldest and more cited article is Nicholson et al., 1999 (7). In this article the term metabolomics was coined as "the quantitative measurement of the dynamic multiparametric metabolic response of living systems to pathophysiological stimuli or genetic modification" and the aim of metabonomics studies "...However, metabonomics deals with detecting, identifying, quantitating and cataloguing the history of time-related metabolic changes in an integrated biological system rather than the individual cell" and finally the authors suggest that computer science and biochemistry are areas where metabonomics field is expected to grow.
Viant et al., (2002) made a research about whiter-ing syndrome in red abalone, they used HNMR and pattern recognition to identify novel bio-markers of these disease, finding that homarines, adenylates and aromatic amino acids showed the highest impact. They argue that one of the challenges of metabolomic studies should be focused on the management of biological variability.
Robertson (2005) published a review about metabolomics applied to toxicological studies, it is one of the most cited article because he starts the review with a description and comparison about metabolomics and metabonomics' terms and made a very interesting analogies, and then a platform (NMR and mass spectrometry) comparison is made emphasizing the strengths and weaknesses of each technique. Then the chemometric challenges that must be faced, the state of technology in 2005 and finally the application of this particular area of toxicology which argues that in vitro toxicity tests based on metabolomic studies have great potential and there is little literature about it. From biomarker identification perspective author found that non supervised techniques such as principal component analysis (PCA) was the most used analysis; up to date PCA is used to visualize data, for outliers' identification and to reduce the dimension of the variables and find the latent variables to be used in a further model or statistic assay. Furthermore, the author argues that understand the mechanism of action should be the most important goal in a me-tabolomics applied to toxicology studies in order to give added value to biomarker discovery. These final idea was then clarified by Xia et al., (2013) where they explain that not always biomarkers help to explain biology phenomenon. Rather they are designed to discriminate different groups of organisms without regard to biological interpretation. Moreover pathway analysis give a biological understanding and may help to develop a treatment (18).
Also in classics there are many others reviews about environmental metabolomics (19, 20) they were focused on describing bioavailability though biomarkers in different organisms but especially in earthworms that will be discussed later. Bundy et al., (2004) and Jones et al., (2008) worked with earthworms exposed to metals and polyaromatic hydrocarbon pyrene respectively; both found changes in TCA cycle intermediates (21, 22).
Sample preparation in metabolomics assessments
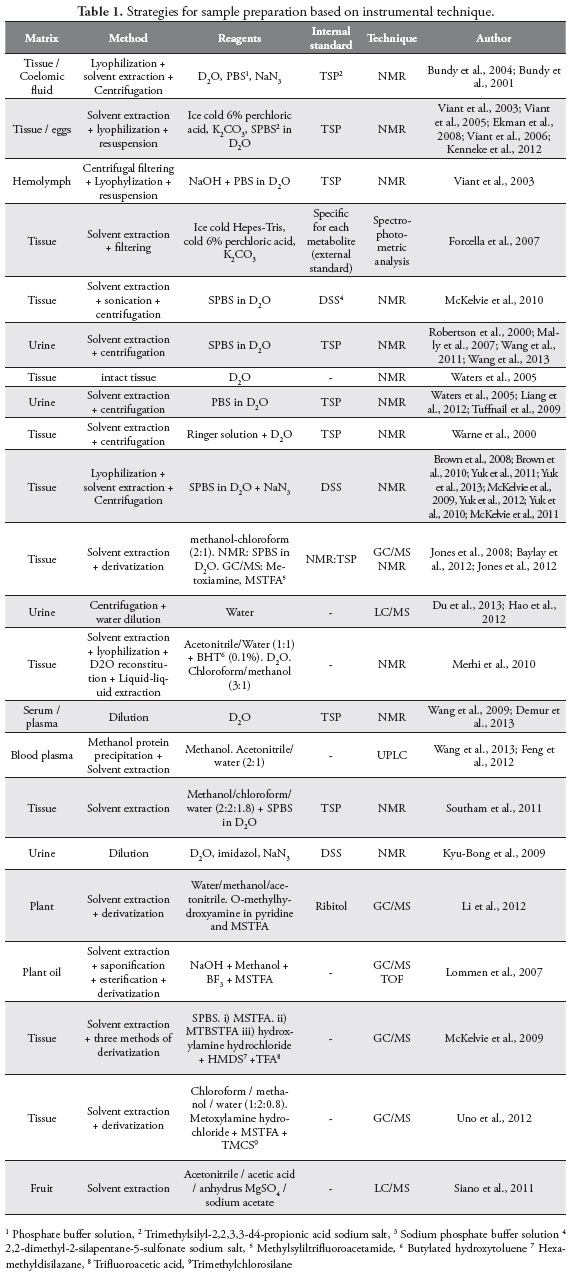
Sample preparation depends primarily on the instrumental technique used in the analysis and secondly on the samples chemical features. Brown et al. (2008) compared different solvent, time of depuration and lyophilization in order to find the best procedure of earthworms sample preparation for NMR analysis (23). They analyzed six different solvents (D2O-based phosphate buffer, acetonitrile-d3, benzene-d6, chloroformd, methanol-d4, DMSO-d6) found that D2O-based phosphate buffer has an excellent reproducibility, highest concentration and greatest variety of metabolites, providing the most detailed metabolic profile, also less variation in the results after longer depuration time and, they found few differences in the data of the earthworms homogenized before lyophilization and those lyophilized individually. Moreover, McKelvie et al. (2009) (24) compared three different methodologies ofsample derivatization for GC/MS analysis from the literature. (i) MSTFA, ii) MTBSTFA, iii) hydroxylamine hydrochloride, HMDS, TFA). They chose the third one because it allow simultaneous analysis of sugars, sugar alcohols, amino acids and yielded the greatest numbers of metabolites in their samples. Table 1. shows the methodologies used on sample preparation based on different instrumental techniques.
Instrumental analysis
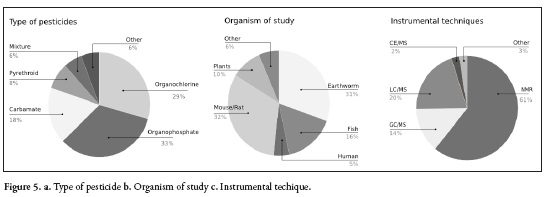
Regarding the instrumental techniques used for metabolomic and pesticide studies, it was found that most studies have been conducted through NMR analyses (25). However, in the last five years, chromatography-mass spectrometry has been increasing used because of its sensitivity. Figure 5c includes the chart that indicate the techniques used in the articles selected for this review.
NMR has been widely applied in metabolomics and pesticides studies. It is a robust method and can directly be used to identify and quantify metabolites without calibration curves because the intensity of the signal is proportional to the molar concentration of the compound (26). Other strengths of this method is the simple sample preparation, it is also a very reproducible method and it is a nondestructive technique.
In pesticide exposure has been mostly used on controlled experiments, possibly because one of the most criticized aspects of this technique is the low sensitivity. However, nowadays this feature has been improved.
The parameters used to acquire and process NMR experiment vary according to the features of the instrument and if it is one or two dimensional spectroscopy. In this review we found that the most used parameters for one dimensional NMR analysis was 500 MHz of frequency, 60° flip angle, 8 KHz of spectral width, 16 to 32K of time domain point, 0.5 Hz of Exponential line broadening and manual baseline correction.
In the other hand, mass spectrometry is becoming the most popular technique used in metabolomics studies because of its availability and versatility. The most common ionization mode found in this review is electrospray ionization (ESI) and the most frequent combination found is LC-MS TOF, as it provides a very good sensitivity and selectivity for a wide range of compounds. However, metabolite identification for LC/MS is still expensive and time consuming because the spectral and retention time libraries for LC/MS is limited and should be built in each laboratory. Furthermore, we found that in many LC/MS based metabolomic studies other experiments was also required in order to identify certain biomarkers. In this vein GC/MS and NMR have an advantage as they can use open access libraries and repositories.
Data Processing
Once the NMR or MS data is acquired, data processing is one of the crucial steps in metabolomics assessments. Baseline correction, water signal suppression, peak peaking, peak deconvolution, isomers and adducts detection, peak alignment and peak annotation are required to obtain the list of metabolites (27). For this, nowadays there is a big list of open source software and web based tools for data processing and it is a growing community. Nevertheless, in this review we found that the data processing was mostly run in commercial software. In the review of Katajamaa et al., (2007) they present two tables of open source and commercial platforms for data processing. A trend of the data processing software is to integrate tools for data analysis and in many articles we found that the software used for data processing was used for data analysis as well (28).
Data analysis
Metabolomics data sets used to have hundreds through thousands variables, and more explanatory variables than number of observations and the metabolites are highly correlated. For this, multi-variate statistics analysis helps to find the mining of metabolomics data. Most of the analysis starts with a non-supervised methodology like Principal Component Analysis (PCA) to describe and visualize the variation in the data set. Is the most used analysis for metabolomics data because it allow reduce the dimension of the variables (metabolites), select the variables that show more variation between treatments or experimental groups, allow to spatially separate the experimental groups and find outliers. Subsequently different analysis is performed according with the research question and experimental design. In this review we found mostly analysis for biomarker selection and most recently followed by pathway analysis.
PLS and OPLS are commonly used for classification and biomarker selection (29) followed by the validation of the models through cross-validation and permutation test. Recently Xia et al., (2015) published the necessity to report the area under receiver operating characteristic and the fold change in biomarker discovery, as this two analysis are widely considered to be the most objective and statistically valid for biomarker discovery (30).
Pathway analysis is used to give a biological meaning of the metabolomics data, linking the metabolites found relevant in the multivariate analysis with the different biological pathways available in public databases like Kegg (http://www.genome.jp/kegg/) or HMDB (http://www.hmdb.ca/).
The analysis of biochemical pathways in metabolo-mics studies have emerged in the last decade in order to understand the data from the perspective of systems biology. The principle is based on two types of analysis, the first is the "Enrichment Set Metabolite analysis (MSEA)" which comes from the analysis of genomic data called "Gen set enrichment analysis (GSEA)" where the principal idea is to investigate enrichment predefined groups of genes instead of studying them individually. In this sense, MSEA focuses on identifying groups of metabolites in biochemical pathways (according to the organism) and according to the list of metabolites obtained in the previous statistical analysis. This comparison can be done by algorithms "overrepresentation analysis (ORA)," "single sample profiling (SSP)" and "quantitative enrichment analysis (QEA)" (31); which integrate statistical analysis and Fisher's test, hypergeometric test, test and analysis overall global covariance (ANCOVA Global).
The second type of analysis is the measure of centrality of metabolites in the network topology. Centrality is a local quantitative measurement of the relative position of a node relative to other nodes and is a measure of the relative importance of a node in a network. Because the biochemical pathways are directed graphs centrality measures used to determine the most important are intermediation (betweenness centrality) or output level (outdegree centrality) (32). These algorithms are integrated and automated cross-platform open access as Metaboanalyst (http://www.metaboana-lyst.ca/) MetPA (http://metpa.metabolomics.ca/ MetPA/) and MSEA (http://www.msea.ca/)
Structural articles
Articles with highest betweeness are those who are linking the classics with the recent with a high rate of centrality, those have an important number of citation and also cite articles with the greatest in degree. These articles are very connected into the network.
A trend to use subletal doses was identified, because metabolomic studies help determine the state of an organism even when there is no observed adverse effect which is one of the added values for studies in toxicology.
McKelvie et al., (2009) evaluated the metabolic response of earthworms exposed to a subletal dose of DDT and endosulfan and they used H-NMR and GC/MS to confirm biomarkers, finding that maltose, alanine and leucine were the metabolites with the greatest variation in the PCA, but only al-anine could be confirmed though both techniques and were present in the two pesticides (24).
Liang et al., 2012 assessed propoxur exposure of rats at different doses above and below the NOAEL, then profiled the metabolites by NMR in serum and urine and the results were associated with different toxic mechanisms found that there was increase in intermediary metabolites of tricarboxylic acid cycle indicating the influence of propoxur in energy metabolism; increased acetate urine indicate that was inhibited acetyl-CoA and that the energy status in liver was altered. Hydroxybutyrate lift is one of the products of fatty acid oxidation in the liver and is key to liver malfunction (33).
Wang et al., 2009 evaluated the subchronic exposure to chlorpyrifos and carbaryl in rats' serum finding that sub-chronic exposure to chlorpyrifos alone or in combination with carbaryl could cause a disturbance in energy and fatty acid metabolism in rat liver mitochondria (34).
Recent articles
Articles with greater out-degree are these recent articles that are very connected within the network. In this section the tendency of the metabolomics studies of pesticide exposure was correlate the main metabolites that showed the highest variation with pathway analysis, in order to present a biological and physiological interpretation.
Xu et al., (2015), evaluated the toxic effects of butachlor on goldfish through a H-NMR based metabolomics approach, monitoring different organs (gill, brain, liver and kidney); they found that the increase of glutathione peroxidase and methane dicarboxylic aldehyde level reflected the disturbance in antioxidative system in the intoxicated gold fish and the increases in lactate dehydrogenase and creatinine suggest liver and kidney injuries (35). Moser et al., 2015 identified potential biomarkers in rat serum exposed to a different chemical class of pesticides (permethrin, deltamethrin, imidacloprid, carbaryl, triadimefon, fipronil), Finding mayor differences in acylcarni-tines, amino acids and glycerophospholipids (36). Olsvik et al., (2015) presented the effect of vitamin E on fatty acid accumulation and carbohydrates metabolism and how vitamin E supplementation can modify chlorpyrifos toxicity in salmon liver cells (37). Bath et al., (2015) presents the effect of the herbicide 2,4-dichlorophenoxyacetic acid (2,4-D) on the nitrogen fixing bacteria in order to determine the effect on specific metabolic pathways and phenotypic changes in sublethal exposure. They found that 2,4-D causes oxidative stress due to decrease in pyrimidine metabolites such as thymidine, uridine 5-phosphate and uracil which are indicative of DNA damage resulting from oxidative stress. Exposure to 2,4-D alter the peptidoglycan and produces intracellular proteins that damage could be determined by the increases of amino acids indicating protein denaturation and proteolysis. Disruption of energy metabolism is correlated with adaptation to stress caused by 2,4-D, which was seen by the increase in related metabolites of the tricarboxylic acid pathway as citrate, phosphoenolpyruvate, glucose 6 phosphate, oxoglutarate and succinate. Protein acetylation and myo-inositol are potential biomarkers of stress adaptation, this was observed with increasing levels of acetyl-lysine and increased inositol phosphate (38).
Wang et al., (2014) evaluated the combined effect of different organophosphorus pesticides in a sub chronic dosage, finding that the exposure to low dose either alone or in combination lead to alteration of liver glucose, fat and protein metabolism, energy metabolism and oxidative balance (1).
Zuluaga et al., (2016) evaluated the effect of different organochlorine pesticides (endoslulfan, aldrin, DDT, lindane and the mixture of all of them) in liver cell culture, finding that each exposure show different metabolic response despite to be organochlorines. Nevertheless, all of them showed sugars altered, suggesting the high requirement of energy to expel the xenobiotic from the cell (94).
Metabolomics and pesticides from different perspectives based on network analysis
The citation network showed different communities showed in figure 3. In each community we identified a trend that gave rise to discuss different perspectives on the role of metabolomics in the study of pesticide exposure.
Modes and mechanisms of action
In the narrative review by Aliferis and Jabaji (2011), a clear distinction is made between the mode of action and mechanism of action. The mode of action describes specific biochemical interactions that are mainly attributed to bioactivity, whereas the mechanism of action refers to the description of all of the biochemical events leading to toxicity of a molecule (39).
Organochlorine, Organophosphate, Carbamate and Pyrethroid are the most investigated type of pesticides found in this review, as those represent more than 80%, Figure 5a, similar conclusion were found about their mode and mechanism of action bellow compiled.
For Organochlorine pesticides is reported the inhibition of neurotransmitter GABA that produces a decrease uptake of chlorine ions in neurons, resulting in partial repolarization and leads convulsions and muscle contractions in organisms (40) (44). For Organophosphate and Carbamate have similar behavior. Inhibition of Acetylcholinesterase (AChE) leads to accumulation of the neurotransmitter acetylcholine in the sympathetic and parasympathetic fibers and in neuromuscular junctions within the vertebrate central nervous system, disturbing transmission across cholinergic synapse and resulting in a variety of neurotoxic effects. Also, chronic exposure could induce oxidative stress leading to lipid peroxidation, DNA damage and protein oxidation (45, 53). For Pyrethroids, membrane sodium channels are interfered, causing lower amplitude of the action potentials or total blockage of neural activity. It produces neuroexcitatory effects in the brain and salivation in mammals, a hallmark of the choreoathetosis with salivation intoxication syndrome. Pyrethroids also act on some calcium channel, an effect that may contribute to the release of neurotransmitters (33, 47, 54)
Aliferis and Jabaji (2011) also compiled more than 85 different modes of action, described by the type of pesticide and their active compound. However, many of those modes of action were described before metabolomics studies started and confirmed later with metabolite profiling assays. For this reason the authors propose to integrate other "omics" to provide new insights into the function and regulation of metabolic networks (39).
Toxicological and biological monitoring
Metabolomics studies on biological or toxico-logical monitoring allows for the oversight of a live system in real time. For this reason, it is possible to study small changes or modifications to metabolism (55). Moreover, the objective of metabolomics as a tool for biological monitoring is to extract latent biochemical information of diagnostic or prognostic value that reflects actual biological events (55). Therefore, detecting such variations can be performed earlier than typical end measures, such as growth inhibition and mortality (25, 56, 57).
To date, metabolomics has had perhaps the greatest impact on the field of toxicology, especially preclinical toxicology, and it is now recognized as an independent technique and widely used for identifying toxicity in target organs and assessing toxicity of chemical agent candidates (58) but the main limitation of metabolomics in toxicology monitoring has been recognizing and separating toxicological effects from physiological effects and determining the adaptive effect of the toxic response because of the difficulty in recovering biological information (55).
Rats and mouse models are commonly used to evaluate toxicological assay and in this review we found them the largest organisms studied (Figure 5b).
Environmental metabolomics and bioavailability
Environmental metabolomics studies are focused on the metabolic changes that occur in organisms in response to an exposure event or environmental stress (20, 59, 60) and bioavailability studies using metabolomics are focused on monitoring biochemical markers that explain the presence or absence of xenobiotics in the environment. In many cases, these compounds undergo transformation and degradation, which is why they may not be found in their original form or in their environmental matrix as soil, water, plants, air and so on. However, it is possible to identify these compounds by metabolites that are expressed in the organism of study (61).
In this review we found that environmental metabolomics has been mostly applied in the study of soil health, pesticide in soil and pesticide residues using earthworm models (62, 66) and metabolomics as a tool to study plant defenses and profiling (67, 69). Thirty-one percent of the articles reviewed used the worm as an organism of study and 10% in plants (Figure 5b).
Metabolism
Metabolism of xenobiotic has been described as a targeted analysis that aims to identify biochemical reactions and quantify the compound or sub-products resulting in the cascade of xenobiotic catabolism or metabolic trajectories (70). It also analyzes changes in the concentration of metabolites that are precursors and products of enzyme activity and then associates these changes with biological functions and/or regulation (71), mostly using graph theory to map the pathways and time-series analysis for the data set. To perform these studies (lately called "fluxomic analysis" (72)) labeled compound are mostly used.
In this literature review we found that energy metabolism is the highest reported. The decrease in sugars is most likely a result of high ATP production. This extra energy produced before a stressful event is mainly a result of the action of cytochrome p450 in the removal of the xenobiotic (19, 20, 44, 50, 59, 70, 71, 73, 75). Similarly, the increase in glycine and alanine is related to the cytoprotective effect they exert with cytochrome p450 (23, 58, 59, 76, 82). And the stereoselectivity of the xenobiotics degradation (83, 84).
Dose response
Chronic exposure to xenobiotics or pesticides often occurs at the no-observed-adverse-effect-level (NOAEL), which is the highest level of continuous exposure to a chemical that does not cause a significant adverse effect to the morphology, biochemistry, functional capacity, growth or lifespan of individuals of a species (85). For this reason, conventional toxicological methods are not always suitable for assessments of toxicological risk and evaluations of pesticide combinations; especially at lower concentrations (58) but it has been the role of metabolomics in dose response assays as it is suitable to detect systemic toxic effects at an earlier stage compared to clinical chemistry (86). NMR and Mass spectrometry platforms are the most common used (Figure 5c). NMR is longer being used, but studies with MS are growing, as this is more sensitive.
Biomarkers
This area of metabolomics has been further explored, and it seeks to separate a large number of metabolites that reveal a direct involvement with the observed phenomenon by minimizing the universe of metabolites that are involved in a reaction but are sufficient to explain it. For this type of study, statistical methods are used that minimize the number of variables and explain differences between systems, treatments or organisms.
Pattern recognition analysis is one of the most frequently used methods for analyzing NMR data (55, 87) because it allows for the extraction of signal characteristics by segmenting the spectrum and includes multivariate statistical methodologies, such as principal component analysis (PCA), partial-least squares (PLS) regression, hierarchical clustering analysis (HCA) and dendrograms (88) as we discussed above in section of data analysis.
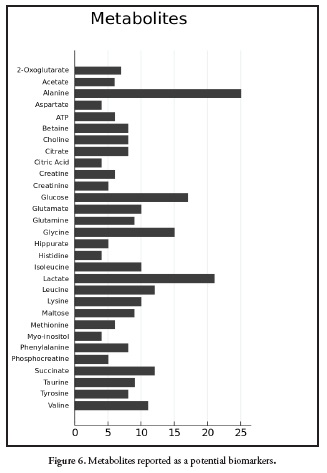
Figure 6. Shows the frequency of metabolites reported in the literature review. Alanine, glucose, glycine and lactate have been reported as possible biomarkers of pesticides exposure. However, those metabolites are also associated with hypoxia in aquatic organisms (40) and starvation (20, 22, 89, 90). For this reason metabolomics has more strength in controlled systems than in epidemiological studies. Also, the study of unknown metabolites is a growing trend for the annotation of biomarkers (91, 92); actually, graph theory is used to find association between mass spectras, linking some knowns metabolites with unknowns to give an approach to the structure of the unknown compound, for this, the tool MetaMap R allow to create the network connecting the metabolites according to the mass spectra (93).
Finally, Based on the literature reviewed, this is the first systematic review that has applied graph theory to analyze the references in studies of me-tabolomics and metabonomics. This analysis has allowed the articles to be viewed as a large network that combines the different interactions among the studies in a wide period of time and makes it possible to identify different perspectives or branches in the study of metabonomics and metabolomics. Moreover, the analysis of the references through graph theory represents a cross-sectional methodology for any search of specific databases because the links retrieve references that are cited in the articles from the search results regardless of the databases to which the journals belong. Use graph theory to select the articles helps to make the review process more efficient and effective.
References
1. Z.-J. Wang, Y. Gao, Y.-L. Hou, C. Zhang, S.-J. Yu, Q. Bian, Z.-M. Li, and W.-G. Zhao, "Design, synthesis, and fungicidal evaluation of a series of novel 5-methyl-1H-1,2,3-trizole-4-carboxyl amide and ester analogues.," Eur. J. Med. Chem., vol. 86, pp. 87-94, Oct. 2014. [ Links ]
2. M. Gassel, C. Wolf, S. Noack, H. Williams, and T. Ilg, "The novel isoxazoline ectoparasiticide fluralaner: selective inhibition of arthropod y-aminobutyric acid- and L-glutamate-gated chloride channels and insecticidal/acaricidal activity.," Insect Biochem. Mol. Biol., vol. 45, pp. 111-24, Feb. 2014. [ Links ]
3. Y.-D. Li, W.-T. Mao, Z.-J. Fan, J.-J. Li, Z. Fang, X.-T. Ji, X.-W. Hua, G.-N. Zong, F.-Y. Li, C.-L. Liu, and J.-H. Yu, "Synthesis and biological evaluation of novel 1,2,4-triazole containing 1,2,3-thiadiazole derivatives," Chinese Chem. Lett., vol. 24, no. 12, pp. 1134-1136, Dec. 2013. [ Links ]
4. H.-N. Guan, D.-F. Chi, J. Yu, and S.-Y. Zhang, "Novel photodegradable insecticide W/TiO(2)/Avermectin nanocompos-ites obtained by polyelectrolytes assembly.," Colloids Surf. B. Biointerfaces, vol. 83, no. 1, pp. 148-54, Mar. 2011. [ Links ]
5. M. P. Longnecker, M. S. Wolff, B. C. Gladen, J. W. Brock, P. Grandjean, J. L. Jacobson, S. a. Korrick, W. J. Rogan, N. Weisglas-Kuperus, I. Hertz-Picciotto, P. Ayotte, P. Stewart, G. Winneke, M. J. Charles, S. W. Jacobson, É. Dewailly, E. R. Boersma, L. M. Altshul, B. Heinzow, J. J. Pagano, and A. a. Jensen, "Comparison of Polychlorinated Biphenyl Levels across Studies of Human Neurodevelopment," Environ. Health Perspect., vol. 111, no. 1, pp. 65-70, Dec. 2003. [ Links ]
6. G. Ding and Y. Bao, "Revisiting pesticide exposure and children's health: Focus on China," Sci. Total Environ., vol. 472, pp. 289-295, 2014. [ Links ]
7. J. K. Nicholson, J. Lindon, and E. Holmes, "'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data," Xenobiotica, vol. 29, no. 11, pp. 1181-1189, 1999. [ Links ]
8. R. N. Trethewey, A. J. Krotzky, and L. Willmitzert, "Commentary Metabolic profiling : a Rosetta Stone for genomics ?," Curr. Opin. P'lant Biol., vol. 2, pp. 83-85, 1999. [ Links ]
9. O. Fiehn, "Metabolomics - The link between genotypes and phenotypes," Plant Mol. Biol, vol. 48, no. 1-2, pp. 155-171, 2002. [ Links ]
10. S. Robledo-giraldo, J. I. Zuluaga-giraldo, and N. D. Duque-Méndez, "Diffusion of Products Through Social Networks :," Respuestas, vol. 18, no. 2, pp. 28-42, 2013. [ Links ]
11. S. Robledo-Giraldo, G. A. Osorio-Zuluaga, and C. López-Es-pinosa, "Networking en pequeña empresa: una revisión bibliográfica utilizando la teoría de grafos," Rev. Vinculos, vol. 11, no. 2, pp. 6-16, 2014. [ Links ]
12. M. Bastian, S. Heymann, and M. Jacomy, "Gephi: An open source software for exploring and manipulating networks. BT - International AAAI Conference on Weblogs and Social," pp. 361-362, 2009. [ Links ]
13. W. D. Wallis, A Beginner's Guide to Graph Theory. Birkhäuser Boston, 2007. [ Links ]
14. B. Bollobás, "The evolution of random graphs," Trans. Am. Math. Soc., vol. 286, no. 1, pp. 257-257, 1984. [ Links ]
15. L. C. Freeman, "A Set of Measures of Centrality Based on Betweenness," Sociometry, vol. 40, no. 1, pp. 35-41, 1977. [ Links ]
16. C. Ni, C. R. Sugimoto, and J. Jiang, "Degree , Closeness , and Betweenness: Application of group centrality measurements to explore macro-disciplinary evolution diachronically," in 13th Meetings of International Society for Scientometrics and Informetrics (ISSI), Durban, South Africa, 2011, pp. 1-13. [ Links ]
17. V. D. Blondel, J. Guillaume, and E. Lefebvre, "Fast unfolding of communities in large networks," J. Stat. Mech. Theory Exp., vol. 10, p. 1000, 2008. [ Links ]
18. J. Xia, D. I. Broadhurst, M. Wilson, and D. S. Wishart, "Translational biomarker discovery in clinical metabolomics: an introductory tutorial," Metabolomics, vol. 9, no. 2, pp. 280-299, 2013. [ Links ]
19. J. G. Bundy, M. P. Davey, and M. R. Viant, "Environmental metabolomics: a critical review and future perspectives," Metabolomics, vol. 5, no. 1, pp. 3-21, Dec. 2009. [ Links ]
20. M. J. Simpson and J. R. McKelvie, "Environmental metabo-lomics: new insights into earthworm ecotoxicity and contaminant bioavailability in soil.," Anal. Bioanal. Chem., vol. 394, no. 1, pp. 137-49, May 2009. [ Links ]
21. J. G. Bundy, D. J. Spurgeon, C. Svendsen, P. K. Hankard, J. M. Weeks, D. Osborn, J. C. Lindon, and J. K. Nicholson, "Environmental metabonomics: applying combination bio-marker analysis in earthworms at a metal contaminated site.," Ecotoxicology, vol. 13, no. 8, pp. 797-806, Nov. 2004. [ Links ]
22. O. a H. Jones, D. J. Spurgeon, C. Svendsen, and J. L. Griffin, "A metabolomics based approach to assessing the toxicity of the polyaromatic hydrocarbon pyrene to the earthworm Lumbricus rubellus.," Chemosphere, vol. 71, no. 3, pp. 601-9, Mar. 2008. [ Links ]
23. S. a E. Brown, A. J. Simpson, and M. J. Simpson, "Evaluation of sample preparation methods for nuclear magnetic resonance metabolic profiling studies with Eisenia fetida.," Environ. Toxicol. Chem., vol. 27, no. 4, pp. 828-36, Apr. 2008. [ Links ]
24. J. R. McKelvie, J. Yuk, Y. Xu, A. J. Simpson, and M. J. Simpson, "1H NMR and GC/MS metabolomics of earthworm responses to sub-lethal DDT and endosulfan exposure," Metabolomics, vol. 5, no. 1, pp. 84-94, Aug. 2009. [ Links ]
25. S. Uno, A. Shintoyo, E. Kokushi, M. Yamamoto, K. Nakayama, and J. Koyama, "Gas chromatography-mass spectrometry for metabolite profiling of Japanese medaka (Oryzias latipes) juveniles exposed to malathion.," Environ. Sci. Pollut. Res. Int., vol. 19, no. 7, pp. 2595-605, Aug. 2012. [ Links ]
26. A. Lubbe, K. Ali, R. Verporte, and Y. H. Choi, "9. NMR- Based Metabolomics Analysis," in Metabolomics in practice: Successful Strategies to Generate and Analyze Metabolic Data, M. Lammerhofer and W. Weckwerth, Eds. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA, 2013, pp. 209-234. [ Links ]
27. M. Sugimoto, M. Kawakami, M. Robert, and T. Soga, "Bioin-formatics tools for mass spectroscopy-based metabolomic data processing and analysis," ... Bioinforma., pp. 96-108, 2012. [ Links ]
28. M. Katajamaa and M. Oresic, "Data processing for mass spec-trometry-based metabolomics.," J. Chromatogr. A, vol. 1158, no. 1-2, pp. 318-28, Jul. 2007. [ Links ]
29. E. Szymanska, E. Saccenti, A. K. Smilde, and J. a Westerhuis, "Double-check: validation of diagnostic statistics for PLS-DA models in metabolomics studies.," Metabolomics, vol. 8, no. Suppl 1, pp. 3-16, Jun. 2012. [ Links ]
30. J. Xia, I. V. Sinelnikov, B. Han, and D. S. Wishart, "Metabo-Analyst 3.0--making metabolomics more meaningful," Nucleic Acids Res., pp. 1-7, 2015. [ Links ]
31. J. Xia and D. S. Wishart, "MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data," Nucleic Acids Res., vol. 38, no. Web Server, pp. W71-W77, 2010. [ Links ]
32. T. Aittokallio and B. Schwikowski, "Graph-based methods for analysing networks in cell biology.," Brief. Bioinform., vol. 7, no. 3, pp. 243-55, Sep. 2006. [ Links ]
33. Y.-J. Liang, H.-P. Wang, D.-X. Long, and Y.-J. Wu, "(1)H NMR-based metabonomic profiling of rat serum and urine to characterize the subacute effects of carbamate insecticide propoxur.," Biomarkers, vol. 17, no. 6, pp. 566-74, Sep. 2012. [ Links ]
34. H.-P. Wang, Y.-J. Liang, D.-X. Long, J.-X. Chen, W.-Y. Hou, and Y.-J. Wu, "Metabolic profiles of serum from rats after subchronic exposure to chlorpyrifos and carbaryl.," Chem. Res. Toxicol., vol. 22, no. 6, pp. 1026-33, Jun. 2009. [ Links ]
35. H.-D. Xu, J.-S. Wang, M.-H. Li, Y. Liu, T. Chen, and A.-Q. Jia, "1H NMR based metabolomics approach to study the toxic effects of herbicide butachlor on goldfish (Carassius auratus)," Aquat. Toxicol., vol. 159, pp. 69-80, 2015. [ Links ]
36. V. C. Moser, N. Stewart, D. L. Freeborn, J. Crooks, D. K. MacMillan, J. M. Hedge, C. E. Wood, R. L. McMahen, M. J. Strynar, and D. W. Herr, "Assessment of serum biomarkers in rats after exposure to pesticides of different chemical classes," Toxicol. Appl. Pharmacol., vol. 282, no. 2, pp. 161-174, 2015. [ Links ]
37. P. a. Olsvik, M. H. G. Berntssen, and L. S0fteland, "Modifying Effects of Vitamin E on Chlorpyrifos Toxicity in Atlantic Salmon," PLoS One, vol. 10, no. 3, p. e0119250, 2015. [ Links ]
38. S. V. Bhat, S. C. Booth, S. G. K. McGrath, and T. E. S. Dahms, "Rhizobium leguminosarum bv. viciae 3841 Adapts to 2,4-Dichlorophenoxyacetic Acid with 'Auxin-Like' Morphological Changes, Cell Envelope Remodeling and Upregu-lation of Central Metabolic Pathways," PLoS One, vol. 10, no. 4, p. e0123813, 2015. [ Links ]
39. K. a. Aliferis and S. Jabaji, "Metabolomics - A robust bioan-alytical approach for the discovery of the modes-of-action of pesticides: A review," Pestic. Biochem. Physiol., vol. 100, no. 2, pp. 105-117, Jun. 2011. [ Links ]
40. W. Tuffnail, G. a. Mills, P. Cary, and R. Greenwood, "An environmental 1H NMR metabolomic study of the exposure of the marine mussel Mytilus edulis to atrazine, lindane, hypoxia and starvation," Metabolomics, vol. 5, no. 1, pp. 33-43, Nov. 2008. [ Links ]
41. J. Yuk, M. J. Simpson, and A. J. Simpson, "1-D and 2-DNMR-based metabolomics of earthworms exposed to endo-sulfan and endosulfan sulfate in soil," Environ. Pollut., vol. 175, pp. 35-44, 2013. [ Links ]
42. J. Yuk, M. J. Simpson, and A. J. Simpson, "1-D and 2-D NMR metabolomics of earthworm responses to sub-lethal trifluralin and endosulfan exposure," Environ. Chem., vol. 8, no. 3, p. 281, 2011. [ Links ]
43. X. Zhang, K. D. Oakes, S. Wang, M. R. Servos, S. Cui, J. Pawliszyn, and C. D. Metcalfe, "In vivo sampling of environmental organic contaminants in fish by solid-phase microextraction," TrAC Trends Anal. Chem., vol. 32, no. Table 1, pp. 31-39, Feb. 2012. [ Links ]
44. M. Li, J. Wang, Z. Lu, D. Wei, M. Yang, and L. Kong, "NMR-based metabolomics approach to study the toxicity of lambda-cyhalothrin to goldfish (Carassius auratus).," Aquat. Toxicol., vol. 146, pp. 82-92, Jan. 2013. [ Links ]
45. Q. Huang and H.-Q. Huang, "Alterations of protein profile in zebrafish liver cells exposed to methyl parathion: a membrane proteomics approach.," Chemosphere, vol. 87, no. 1, pp. 68-76, Mar. 2012. [ Links ]
46. A. D. Southam, A. Lange, A. Hines, E. M. Hill, Y. Katsu, T. Iguchi, C. R. Tyler, and M. R. Viant, "Metabolomics reveals target and off-target toxicities of a model organophosphate pesticide to roach (Rutilus rutilus): implications for biomon-itoring.," Environ. Sci. Technol., vol. 45, no. 8, pp. 3759-67, Apr. 2011. [ Links ]
47. H.-P. Wang, Y.-J. Liang, Y.-J. Sun, J.-X. Chen, W.-Y. Hou, D.-X. Long, and Y.-J. Wu, "1H NMR-based metabonomic analysis of the serum and urine of rats following subchronic exposure to dichlorvos, deltamethrin, or a combination of these two pesticides.," Chem. Biol. Interact., vol. 203, no. 3, pp. 588-96, May 2013. [ Links ]
48. S. Uno, A. Shintoyo, E. Kokushi, M. Yamamoto, K. Nakayama, and J. Koyama, "Gas chromatography-mass spectrometry for metabolite profiling of Japanese medaka (Oryzias latipes) juveniles exposed to malathion.," Environ. Sci. Pollut. Res. Int., vol. 19, no. 7, pp. 2595-605, Aug. 2011. [ Links ]
49. M. Forcella, E. Berra, R. Giacchini, B. Rossaro, and P. Parenti, "Increased alanine concentration is associated with exposure to fenitrothion but not carbamates in Chironomus riparius larvae.," Ecotoxicol. Environ. Saf., vol. 66, no. 3, pp. 326-34, Mar. 2007. [ Links ]
50. A. J. Mearns, D. J. Reish, P. S. Oshida, T. Ginn, and M. A. Rempel-Hester, "Effects of Pollution on Marine Organisms," Water Environ. Res., vol. 83, no. 10, pp. 1789-1852, Jan. 2011. [ Links ]
51. N. Sánchez-Avila, J. M. Mata-Granados, J. Ruiz-Jiménez, and M. D. Luque de Castro, "Fast, sensitive and highly discriminant gas chromatography-mass spectrometry method for profiling analysis of fatty acids in serum.," J. Chromatogr. A, vol. 1216, no. 40, pp. 6864-72, Oct. 2009. [ Links ]
52. N. Bonvallot, M. Tremblay-Franco, C. Chevrier, C. Canlet, C. Warembourg, J.-P. Cravedi, and S. Cordier, "Metabolomics tools for describing complex pesticide exposure in pregnant women in Brittany (France).," PLoS One, vol. 8, no. 5, p. e64433, Jan. 2013. [ Links ]
53. Y.-J. Liang, W. HP, Y. L, L. W, and W. YJ., "Metabonomic responses in rat urine following subacute exposure to propoxur.," Int J Toxicol, vol. 31, no. 3, pp. 287-93, 2012. [ Links ]
54. P. Carriquiriborde, D. J. Marino, G. Giachero, E. a Castro, and A. E. Ronco, "Global metabolic response in the bile of pejerrey (Odontesthes bonariensis, Pisces) sublethally exposed to the pyrethroid cypermethrin.," Ecotoxicol. Environ. Saf., vol. 76, no. 2, pp. 46-54, Feb. 2012. [ Links ]
55. J. K. Nicholson, J. Connelly, J. C. Lindon, and E. Holmes, "Metabonomics: a platform for studying drug toxicity and gene function.," Nat. Rev. Drug Discov., vol. 1, no. 2, pp. 153-61, Feb. 2002. [ Links ]
56. P. Fuentealba González, A. Llanos-Rivera, N. Carvajal Baeza, and E. Uribe Pérez, "Xenobiotic-induced changes in the arginase activity of zebrafish (Danio rerio) eleutheroembryo.," Environ. Toxicol. Chem., vol. 30, no. 10, pp. 2285-91, Oct. 2011. [ Links ]
57. O. a H. Jones, S. Murfitt, C. Svendsen, A. Turk, H. Turk, D. J. Spurgeon, L. a Walker, R. F. Shore, S. M. Long, and J. L. Griffin, "Comparisons of metabolic and physiological changes in rats following short term oral dosing with pesticides commonly found in food.," Food Chem. Toxicol., vol. 59, pp. 438-5, Sep. 2013. [ Links ]
58. H.-P. Wang, Y.-J. Liang, Q. Zhang, D.-X. Long, W. Li, L. Li, L. Yang, X.-Z. Yan, and Y.-J. Wu, "Changes in metabolic profiles of urine from rats following chronic exposure to an-ticholinesterase pesticides," Pestic. Biochem. Physiol., vol. 101, no. 3, pp. 232-239, Nov. 2011. [ Links ]
59. J. Yuk, M. J. Simpson, and A. J. Simpson, "1-D and 2-DNMR metabolomics of earthworm responses to sub-lethal trifluralin and endosulfan exposure," Environ. Chem., vol. 8, no. 3, p. 281, 2011. [ Links ]
60. J. Yuk, J. R. McKelvie, M. J. Simpson, M. Spraul, and A. J. Simpson, "Comparison of 1-D and 2-D NMR techniques for screening earthworm responses to sub-lethal endosulfan exposure," Environ. Chem., vol. 7, no. 6, p. 524, 2010. [ Links ]
61. J. R. McKelvie, J. Yuk, Y. Xu, A. J. Simpson, and M. J. Simpson, "1H NMR and GC/MS metabolomics of earthworm responses to sub-lethal DDT and endosulfan exposure," Metabolomics, vol. 5, no. 1, pp. 84-94, Aug. 2008. [ Links ]
62. S. a E. Brown, J. R. McKelvie, A. J. Simpson, and M. J. Simpson, "1H NMR metabolomics of earthworm exposure to sub-lethal concentrations of phenanthrene in soil.," Environ. Pollut., vol. 158, no. 6, pp. 2117-23, Jun. 2010. [ Links ]
63. J. G. Bundy, D. Osborn, J. M. Weeks, J. C. Lindon, and J. K. Nicholson, "An NMR-based metabonomic approach to the investigation of coelomic fluid biochemistry in earthworms under toxic stress.," FEBSLett., vol. 500, no. 1-2, pp. 31-5, Jun. 2001. [ Links ]
64. T. H. Bundy JG, Lenz EM, Bailey NJ, Gavaghan CL, Svendsen C, Spurgeon D, Hankard PK, Osborn D, Weeks JM, Trauger SA, Speir P, Sanders I, Lindon JC, Nicholson JK, "Metabonom-ic assessment of toxicity of 4-fluoroaniline, 3,5-difluoroaniline and 2-fluoro-4-methylaniline to the earthworm Eisenia veneta (Rosa): identification of new endogenous biomarkers.," Env. Toxicol Chem., vol. 21, no. 9, pp. 1966-72, 2002. [ Links ]
65. J. G. Bundy, D. J. Spurgeon, C. Svendsen, P. K. Hankard, J. M. Weeks, D. Osborn, J. C. Lindon, and J. K. Nicholson, "Environmental metabonomics: applying combination bio-marker analysis in earthworms at a metal contaminated site.," Ecotoxicology, vol. 13, no. 8, pp. 797-806, Nov. 2004. [ Links ]
66. J. G. Bundy, H. C. Keun, J. K. Sidhu, D. J. Spurgeon, C. Svendsen, P. Kille, and A. J. Morgan, "Metabolic Profile Biomarkers of Metal Contamination in a Sentinel Terrestrial Species Are Applicable Across Multiple Sites," Environ. Sci. Technol., vol. 41, no. 12, pp. 4458-4464, 2007. [ Links ]
67. M. Mirnezhad, R. R. Romero-González, K. a Leiss, Y. H. Choi, R. Verpoorte, and P. G. L. Klinkhamer, "Metabolomic analysis of host plant resistance to thrips in wild and cultivated tomatoes.," Phytochem. Anal., vol. 21, no. 1, pp. 110-7, 2010. [ Links ]
68. G. G. Siano, I. S. Pérez, M. D. G. García, M. M. Galera, and H. C. Goicoechea, "Multivariate curve resolution modeling of liquid chromatography-mass spectrometry data in a comparative study of the different endogenous metabolites behavior in two tomato cultivars treated with carbofuran pesticide.," Talanta, vol. 85, no. 1, pp. 264-75, Jul. 2011. [ Links ]
69. I. J. Colquhoun, "Use of NMR for metabolic profiling in plant systems," J. Pestic. Sci., vol. 32, no. 3, pp. 200-212, 2007. [ Links ]
70. M. R. Viant, C. a Pincetich, and R. S. Tjeerdema, "Metabolic effects of dinoseb, diazinon and esfenvalerate in eyed eggs and alevins of Chinook salmon (Oncorhynchus tshawytscha) determined by 1H NMR metabolomics.," Aquat. Toxicol., vol. 77, no. 4, pp. 359-71, May 2006. [ Links ]
71. C. Y. Lin, M. R. Viant, and R. S. TJeerdema, "Metabolomics : Methodologies and applications in the environmental sciences," J. Pestic. Sci., vol. 31, no. 3, pp. 245-251, 2006. [ Links ]
72. K. Sanford, P. Soucaille, G. Whited, and G. Chotani, "Genomics to fluxomics and physiomics - pathway engineering," Curr. Opin. Microbiol., vol. 5, no. 3, pp. 318-322, Jun. 2002. [ Links ]
73. M. G. Miller, "Environmental metabolomics: a SWOT analysis (strengths, weaknesses, opportunities, and threats).," J. Proteome Res., vol. 6, no. 2, pp. 540-5, Feb. 2007. [ Links ]
74. M. Merhi, C. Demur, C. Racaud-Sultan, J. Bertrand, C. Can-let, F. B. Y. Estrada, and L. Gamet-Payrastre, "Gender-linked haematopoietic and metabolic disturbances induced by a pesticide mixture administered at low dose to mice.," Toxicology, vol. 267, no. 1-3, pp. 80-90, Jan. 2010. [ Links ]
75. N. J. Waters, C. J. Waterfield, R. D. Farrant, E. Holmes, and J. K. Nicholson, "Metabonomic deconvolution of embedded toxicity: application to thioacetamide hepato- and nephrotoxicity.," Chem. Res. Toxicol., vol. 18, no. 4, pp. 639-54, Apr. 2005. [ Links ]
76. K.-B. Kim, S. H. Kim, S. Y. Um, M. W. Chung, J. S. Oh, S.-C. Jung, T. S. Kim, H. J. Moon, S. Y. Han, H. Y. Oh, B. M. Lee, and K. H. Choi, "Metabolomics approach to risk assessment: methoxyclor exposure in rats.," J. Toxicol. Environ. Health. A, vol. 72, no. 21-22, pp. 1352-68, Jan. 2009. [ Links ]
77. J. R. McKelvie, J. Yuk, Y. Xu, A. J. Simpson, and M. J. Simpson, "1H NMR and GC/MS metabolomics of earthworm responses to sub-lethal DDT and endosulfan exposure," Metabolomics, vol. 5, no. 1, pp. 84-94, Aug. 2009. [ Links ]
78. M. A. Warne, E. M. Lenz, D. Osborn, and J. M. Weeks, "An NMR-based metabonomic investigation of the toxic effects of 3-trifluoromethyl-aniline on the earth worm Eisenia veneta," Biomarkers, vol. 5, no. 1, pp. 56-72, 2000. [ Links ]
79. H.-P. Wang, Y.-J. Liang, Y.-J. Sun, J.-X. Chen, W.-Y. Hou, D.-X. Long, and Y.-J. Wu, "1H NMR-based metabonomic analysis of the serum and urine of rats following subchronic exposure to dichlorvos, deltamethrin, or a combination of these two pesticides.," Chem. Biol. Interact., vol. 203, no. 3, pp. 588-96, May 2013. [ Links ]
80. C. Demur, B. Métais, C. Canlet, M. Tremblay-Franco, R. Gautier, F. Blas-Y-Estrada, C. Sommer, and L. Gamet-Payrastre, "Dietary exposure to a low dose of pesticides alone or as a mixture: the biological metabolic fingerprint and impact on hematopoiesis.," Toxicology, vol. 308, pp. 74–87, Jun. 2013. [ Links ]
81. O. a H. Jones, S. C. Swain, C. Svendsen, J. L. Griffin, S. R. Sturzenbaum, and D. J. Spurgeon, "Potential new method of mixture effects testing using metabolomics and Caenorhabditis elegans.," J. Proteome Res., vol. 11, no. 2, pp. 1446–53, Feb. 2012. [ Links ]
82. M. K. R. Mudiam, R. Ch, and P. N. Saxena, "Gas chromatography- mass spectrometry based metabolomic approach for optimization and toxicity evaluation of earthworm sub-lethal responses to carbofuran.," PLoS One, vol. 8, no. 12, p. e81077, Jan. 2013. [ Links ]
83. J. F. Kenneke, D. R. Ekman, C. S. Mazur, B. J. Konwick, A. T. Fisk, J. K. Avants, and A. W. Garrison, "Integration of Metabolomics and In Vitro Metabolism Assays for Investigating the Stereoselective Transformation of Triadimefon in Rainbow Trout," Chirality, vol. 192, no. May 2009, pp. 183–192, 2010. [ Links ]
84. Y. Picó and D. Barceló, "The expanding role of LC-MS in analyzing metabolites and degradation products of food contaminants," TrAC Trends Anal. Chem., vol. 27, no. 10, pp. 821–835, Nov. 2008. [ Links ]
85. D. Hao, W. Xu, H. Wang, L. Du, J. Yang, X. Zhao, and C. Sun, "Ecotoxicology and Environmental Safety Metabolomic analysis of the toxic effect of chronic low-dose exposure to acephate on rats using ultra-performance liquid chromatography / mass spectrometry," Ecotoxicol. Environ. Saf., vol. 83, pp. 25–33, 2012. [ Links ]
86. Z. Feng, X. Sun, J. Yang, D. Hao, L. Du, H. Wang, W. Xu, X. Zhao, and C. Sun, "Metabonomics analysis of urine and plasma from rats given long-term and low-dose dimethoate by ultra-performance liquid chromatography-mass spectrometry.," Chem. Biol. Interact., vol. 199, no. 3, pp. 143-53, Sep. 2012. [ Links ]
87. D. G. Robertson, M. D. Reily, R. E. Sigler, D. F. Wells, D. a Paterson, and T. K. Braden, "Metabonomics: evaluation of nuclear magnetic resonance (NMR) and pattern recognition technology for rapid in vivo screening of liver and kidney toxicants.," Toxicol. Sci., vol. 57, no. 2, pp. 326-37, Oct. 2000. [ Links ]
88. K. a. Aliferis and M. Chrysayi-Tokousbalides, "Metabolomics in pesticide research and development: review and future perspectives," Metabolomics, vol. 7, no. 1, pp. 35-53, Aug. 2011. [ Links ]
89. M. R. Viant, E. S. Rosenblum, and R. S. Tjeerdema, "NMR-Based Metabolomics: A Powerful Approach for Characterizing the Effects of Environmental Stressors on Organism Health," Environ. Sci. Technol., vol. 37, no. 21, pp. 4982-89, 2003. [ Links ]
90. J. R. McKelvie, D. M. Wolfe, M. Celejewski, A. J. Simpson, and M. J. Simpson, "Correlations of Eisenia fetida metabolic responses to extractable phenanthrene concentrations through time.," Environ. Pollut., vol. 158, no. 6, pp. 2150-7, Jun. 2010. [ Links ]
91. A. Lommen, G. van der Weg, M. C. van Engelen, G. Bor, L. a P. Hoogenboom, and M. W. F. Nielen, "An untargeted metabolomics approach to contaminant analysis: pinpointing potential unknown compounds.," Anal. Chim. Acta, vol. 584, no. 1, pp. 43-9, Feb. 2007. [ Links ]
92. E. Liotta, R. Gottardo, A. Bertaso, and A. Polettini, "Screening for pharmaco-toxicologically relevant compounds in biosamples using high-resolution mass spectrometry: a 'metabolomic' approach to the discrimination between isomers.," J. Mass Spectrom., vol. 45, no. 3, pp. 261-71, Mar. 2010. [ Links ]
93. D. Grapov, K. Wanichthanarak, and O. Fiehn, "MetaMapR: Pathway Independent Metabolomic Network Analysis In- corporating Unknowns," Bioinforma. Adv. Access, pp. 5-8, 2015. [ Links ]