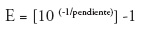Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Nova
versión impresa ISSN 1794-2470
Nova vol.14 no.26 Bogotá jul./dic. 2016
In-house standardization and validation of a multiplex RT-PCR assay for the detection of 13 respiratory viruses
Estandarización interna y validación de un ensayo RT-PRC múltiple para la detección de 13 virus respiratorios
Hernán Vargas1*, Ángela Diaz1, Yamile Celis1, Liliana Díaz1, Sandra Gómez1, Jenny Sánchez1, Carlos Golijow2, Patricia Arce1.
1 Laboratorio de Salud Pública, Dirección de Epidemiología, Análisis y Gestión de Políticas de Salud Colectiva, Subsecretaría de Salud Pública, Secretaría Distrital de Salud; Bogotá, Colombia,
2 IGEVET-Instituto de Genética Veterinaria "Ing. Fernando Dulout" (UNLP-CONICET LA PLATA), Buenos Aires-Argentina
Corresponding: havargas@saludcapital.gov.co
Fecha de recepción: Junio 27 de 2016 - Fecha de aprobación: Octubre 18 de 2016
Abstract
Background. Multiplex real time PCR is increasingly used to diagnose respiratory viruses and has shown to be superior to traditional methods, such as culture and antigen detection. Objective. Standardization and validation of a multiplex real-time PCR assay for the detection of 13 respiratory viruses. Methods. The assay was validated using RNA control targets and comparing results to single-target PCR's. Results. Using RNA controls the multiplex format was found to be as sensitive and specific as the single-target PCRs, and no competition was observed between targets. The efficiencies for most of the reactions were approximately 90%, but a lower efficiency was found for Parainfluenza 2 with a rate of amplification in each cycle of 86.63%. On the other hand, a higher efficiency was observed in respiratory syncytial virus A and respiratory syncytial virus B ((93.07% each). Conclusion: This multiplex RT-PCR format shows an adequate efficiency, demonstrating an excellent sensitivity, specificity and repeatability for all the studied respiratory viruses.
Keywords: Multiplex Real Time PCR, Respiratory virus, Standardization.
Resumen
Antecedentes. PRC múltiple en tiempo real es usada cada vez más para el diagnóstico de virus respiratorios y ha mostrado ser superior a metodos tradicionales, como cultivo y detección de antígeno. Objetivo. Estandarizar y validar una PRC múltiple en tiempo real para la detección de 13 virus respiratorios. Metodos. El ensayo fue realizado usando blanco de RNA control y comparando los resultados a blancos únicos de PCR. Resultados. Usando el RNA control, el formato de multiplex era tan sensible y específico como la PCR. Las eficiencias para la mayoría de las reacciones de aproximadamente el 90%, pero una eficiencia baja fue encontrada para influenza 2 con una tasa de amplificación en cada ciclo de 86.63%. Por otra parte, una mayor eficiencia fue observada en virus sincitial respitario A y B (93, 67% cada uno). Conclusión. Este formato RT-PCR múltiple muestra una adecuada eficiencia, demostrando un excelente especificidad y reproducibilidad para todos los virus respiratorios estudiados.
Palabras Clave: PRC múltiple en tiempo real, virus respiratorio, estandarización.
Introduction
Viral Acute respiratory infections (ARI) remain a major cause of morbidity, mortality and economic losses (1). These infections are often self-limiting. In certain risk groups such as children under 5 years, those over 60 years and immunocompromised people, may show acute clinical symptoms and death (2, 3, 4).
Evaluate the actual impact of infections caused by the most common respiratory viruses, in a qualitative and quantitative way, is and has been a challenge. One reason, is that diagnosis in most countries is based on the use of conventional methods, such as viral culture or immunofluorescence (IF) (1, 5, 6).
Indirect immunofluorescence (IF) is faster but less sensitive than cell culture. Despite being a technique with high specificity, does not have a high sensitivity, leading to false negative results (1). Also, a significant number of samples remain negative, despite clinical suspicion of viral infection (7, 8, 9, 10). Viral culture is still considered as the "gold standard" for respiratory viruses detection, but it is limited by the elapsed time for getting results (up to 14 days) and by the strict conditions for transport and storage of samples in order to conserve virus infectivity (11). For this reason, in the recent years the Polymerase Chain Reaction (PCR) has improved the diagnostic of viral infections, being a powerful tool for the detection and quantification ofRNA or DNA. The real-time PCR is increasingly used in diagnostics due to its high sensitivity and good reproducibility (12, 13). Previous studies have demonstrated the superior sensitivity of multiplex real-time PCR over traditional methods (14, 15).
The objective of the present study was to standardize and validate an in house Multiplex real-time PCR for the detection of 13 respiratory viruses (Influenza A and B, Respiratory Syncytial Virus (RSV) A and B, Adenovirus, Parainfluenza 1, 2 and 3, rhinovirus, coronavirus 229E and coronavirus OC43, metapneumovirus and Human Bocavirus) in respiratory samples, obtained through the sentinel surveillance program from Bogotá, Colombia. Approval by an ethics committee was not requested since this work is part of the program for epidemiological surveillance from the Health Department of Bogota, Colombia.
Materials and Methods
In silico analysis of primers and probes
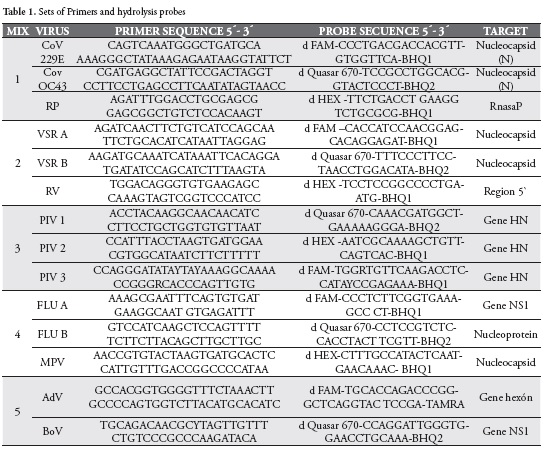
Bioinformatic analysis was performed using Fast-PCR software, version 6.2.96, for all the primers and hydrolysis probes (TaqMan probes), in order to verify their specificity (16, 17, 18). After defining that the primers and probes were adequate for the analysis, the work mixtures were defined, Table 1, and in silico tests were performed, to verify the specificity of such mixtures.
Viral Controls
The nucleic acids controls for Influenza viruses A and B, Parainfluenza 1-3, Respiratory Syncytial Virus and Adenovirus were provided by the National Institute of Health (Colombia). The controls for Rhinovirus, Coronavirus 229E and OC43, Metapneumovirus (cell cultures) and Human Bocavirus (plasmids) were obtained from the Centers for Diseases Control (CDC), Atlanta, USA.
Nucleic Acid Extraction and Real-time PCR
Nucleic acids for Rhinovirus, Coronavirus 229E, Coronavirus OC43, and Metapneumovirus were extracted using QIAamp® Viral RNA Mini Kit (QIAGEN Cat. N°.52906). Nucleic acids were kept frozen at -70°C until use. All PCR reactions (monoplex and multiplex) were performed on a CFX96 thermocycler (BioRad), using exactly the same conditions for all reactions.
Amplifications were performed in a final reaction volume of 25 µl, using 5 µl of control viral nucleic acid and Superscript III Platinum One step q-RT-PCR kit (Invitrogen). Reverse transcription was performed for 30 min at 50°C, the Platinum Taq polymerase was activated at 95°C for 2 min and 44 cycles of PCR performed at 95°C for 15s and 55°C for 30 s.
Standardization
Several trials were conducted in the laboratory. Initially, in monoplex reactions and then in multiplex reactions, testing primers and hydrolysis probes in order to determine the optimum concentration of reactants and the best conditions for amplification.
Threshold cycle (Ct) is indicated in the results section. This value reflects the number of cycles for a particular reaction where the emission intensity of the probe rises above the background noise, crossing the threshold. So that, the lower the Ct value the higher RNA or DNA target is present in the sample.
Measuring of Sensitivity and Specificity
To determine the behavior of the system, in response to variations in the concentration of nucleic acids, an assay of analytical sensitivity was carried out in monoplex and multiplex reactions. These assays were performed in triplicate serial dilutions, in base 10 (1 x 10-1 - 1 x 10-9) from the original control for each virus included in the study. All the controls were quantified to determine the initial concentration.
Specificity assays were performed analyzing potential cross reactions with Bordetella pertussis and Rotavirus. The bacterium Bordetella pertussis can be found in the respiratory tract and Rotavirus was not found because it is a gastrointestinal virus. To perform this evaluation, positive samples for these two pathogens were analyzed using the primers and probes deined for this work.
Efficiency
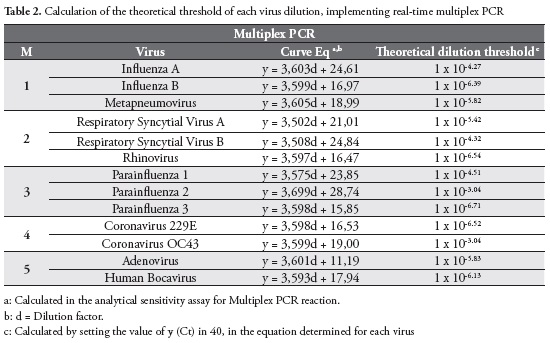
Amplification efficiency of PCR reactions were determined in order to obtain valid comparisons between different samples. To determine the efficiency of amplification in a given primer set, serial dilutions from the original control of each virus included in the study were performed and a standard curve was constructed. The efficiency was calculated according to the following formula:
Repeatability
Repeatability was calculated analyzing three individual aliquots of dilutions with Cts between 20-40. The standard deviation was calculated as an indicator of repeatability in monoplex and multiplex reactions.
Results
Standardization
Standardization of multiplex assays was performed on viral characterized controls, provided by the "Instituto Nacional de Salud" (INS) and the CDC. Initially, monoplex PCRs were performed to ensure the specificity of the primers and probes. In all cases, each virus was detected only for the specific set of primers and probes, being the lowest Ct value for the Adenovirus (10.56) and the highest for respiratory syncytial virus B (24.00). Multiplex PCRs were tested to verify that cross-reactions were avoided and that results were similar to those obtained for monoplex reactions. The lowest Ct value (11.10) was for Adenovirus and the highest for Respiratory Syncytial Virus B (24.93).
The last dilutions showing signal in the Multiplex PCRs were between the ranges of 1 x 10-3 for Parainfluenza 2 and 1 x 10-8 for Adenovirus.
Furthermore, the theoretical dilution threshold for detection of respiratory viruses using the real-time multiplex PCR was calculated, Table 2.
Efficiency
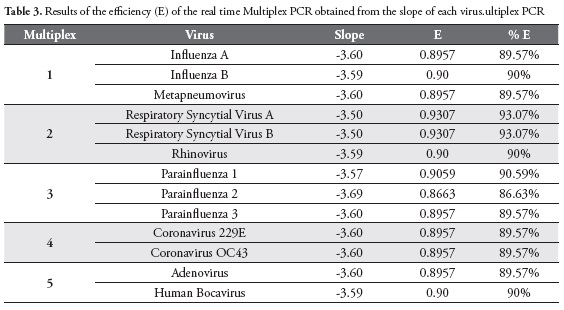
Efficiency was calculated for each virus in Multiplex PCRs reactions, obtaining efficiencies of 93.07% for Respiratory Syncytial Virus A and Respiratory Syncytial Virus B, 90% for influenza B virus, Rhinovirus, and Human Bocavirus. Parainfluenzavirus 2 showed the lowest efficiency (86.63%), Table 3.
Repeatability
Detection of virus RNAs were performed in triplicates. Three independent replicates were performed for both, PCR Monoplex and Multiplex reactions. The runs for Monoplex PCRs showed coefficients of variation between 0.006 - 0.036. For Multiplex PCRs the coefficients of variation were between 0.01 - 0.036. Parainfluenza 1 virus was a case out of range (1.18). These findings indicate a high repeatability for both Monoplex and multiplex PCR assays.
Discussion
Cell culture is still considered as the "gold standard" for the identiication of the most prevalent respiratory viruses. Although this method presents some difficulties, such as instability of cultured cells, maintenance of cell cultures, the long time to obtain results, among others (1, 19, 20). Actually, many methods exist for a quick identification of viral infections. However, the molecular tests have showed excellent performance and may represent an alternative for the diagnostic of routine in the laboratory (1).
This study describes the development of five realtime multiplex PCRs that can run together, for the detection of 13 respiratory viruses.
Tests for sensitivity, specificity and repeatability (from characterized viral controls) were conducted to determine the robustness of the implemented Multiplex real-time PCRs for processing respiratory specimens.
A good sensitivity was found for both the systems, monoplex and multiplex PCRs. The lowest dilution showing signal for the Multiplex system ranges between Ct 37.01 to Ct 40.01 for each virus. Results were very similar for both types of assays, although the Ct values were higher in the Multiplex PCR assays. This situation could be attributed to reaction kinetics, when several primers and probes are in a single mixture.
The specificity tests showed that the primers and probes are specific for the virus included in the study (Influenza A and B, Respiratory Syncytial Virus A and B, Adenovirus, Parainfluenza 1, 2 and 3, Rhinovirus, Coronavirus 229E - OC43; Metapneumovirus and Human Bocavirus ), and not cross reactions were observed when genetic material from Rotavirus and Bordetella pertussis was included.
Theoretically, the number of copies of genetic material doubles each round of PCR, so that for each dilution 1:10 Ct value must have an increase of around 3.32 (21). By plotting the Ct values vs dilution, and calculate the equation of the line, the slope (m) represents this value. This is an important issue because the efficiency of the PCR reaction should be between 90-100% (slope between 3.32 and 3.6) (21). If the efficiency is 100%, the Ct values of 3.32 appeared in each dilution for each cycle. If the slope is less than -3.6, then the efficiency of the PCR begins to decrease. Besides, the value of R2 for a standard curve represents how well the experimental data it the regression line, i.e. how the data are linear. Therefore, ideally the value of R2 should be > 0.99 (21). In this study, the slopes for different viruses for Multiplex PCRs range between 3.5 and 3.69.
The efficiency for PCR amplification is often presented as a percentage, i.e. the percentage of ampliied genetic material in each cycle, inding for most viruses efficiencies around 90%. The lowest efficiency was found for Parainfluenza 2 virus, with a percentage of ampliication of genetic material in each cycle of 86.63%. In contrast, the higher efficiency was found for Respiratory Syncytial Virus A and Respiratory Syncytial Virus B, with a value of 93.07%.
The coefficient of determination (R2), also calculated from the equation of the line, indicates the percentage of variability of the Ct values. In this study, R2 values remained above 0.99, being in most cases 1, indicating a strong correlation between the data.
It is important that the PCR efficiency has adequate values, because for 100% efficiency, the amount of genetic material is doubled in each cycle. With efficiencies of 90% which was obtained by us, the genetic material increase in 1,9 for each cycle. A small difference in PCR efficiency makes a great difference in the quantity of final product, since every time the quantity of obtained product is also lower. This situation forces to perform higher number of cycles to detect a particular amount of genetic material (22).
Performing repeatability tests for the implemented PCR assays is quite important. An excess in variability should be corrected before continuing the process of validation and implementation of the technique. The variation of the detected average value is determined as an indicator of the repeatability. Our Multiplex PCRs have very low values, ranging from 0.01 to 0,036 with a case of anomalous standard deviation of 1.18 for the 10-1 dilution of Parainfluenza virus type 1. Probably, this situation was achieved because the Ct, obtained for the irst replica, was lower than that obtained for the others. An excess of PCR target could generate this kind of anomaly. Except for this discrepancy, the data indicate that PCRs have a good agreement between results for each run, in order to ensure reliable results.
Conclusion
The present study showed a good performance for the multiplex PCR assays, to detect 13 agents tightly associated- to respiratory tract disease. These assays have some advantages, such as not requiring post PCR handling and could be used in quick diagnostic procedures for the detection of respiratory viruses.
Acknowledgements
The authors want to thank Dr. Dean D. Erdman (CDC) and Dr. Jairo Méndez (INS) for kindly making available the viral controls. The authors declare no potential conflicts of interest relevant to this article.
References
1. Salez N., Vabret A., Leruez M., Andreoletti L., Carrat F. et al. (2015). Evaluation of four Commercial Multiplex Molecular Test for the Diagnosis of acute respiratory Infections. Plos One 10 (Vol 6); 1-17. [ Links ]
2. Sanghavi S, Bullota A, Husain S, Rinaldo C. (2012). Clinical evaluation of multiplex real time PCR panels for rapid detection of respiratory viral infections. Journal of medical Virology. (Vol 84); 162-169. [ Links ]
3. Ferkol T. Schraufnagel, D. (2014). The Global Burden of respiratory disease. Ann Am Thorac Soc (Vol 11); 404-406. [ Links ]
4. Watanabe A, Carraro E, Camarago C, Puerari D, Guatura S. et al. (2013). Human adenovirus detection among immunocompetent and immunocompromised patients presenting acute respiratory infection. Revista da Sociedad Brasileira de Medicine Tropical 46 (2); 161-165. [ Links ]
5. Pavia A. (2011). Viral Infections of the lower respiratory tract; Old viruses, new viruses, and the role of diagnosis. Clin Infect Dis. (Vol 4); 284-289. [ Links ]
6. Krause J, Panning M, Hengel H, Henneke P. (2014). The role of multiplex PCR in respiratory tract infections in children. Dtsch Arztebl Int; (111); 639-45. [ Links ]
7. Weissenbacher MC, MM Ávila. Los virus como causa de IRA alta y baja en niños: características generales y diagnóstico, in Infecciones respiratorias en niños 1999, OPS - OMS: http://www.paho.org/Spanish/AD/DPC/CD/aiepi1.htm; 89-105. [ Links ]
8. Liolios L, Jenney A, Spelman D, Kotsimbos T, Catton M. et al. (2001). Comparison of a Multiplex Reverse Transcription-PCR-Enzyme Hybridization Assay with Conventional Viral Culture and Immunofluorescence Techniques for the Detection of Seven Viral Respiratory Pathogens. J Clin Microbiol, 39(8); 2779-2783. [ Links ]
9. Syrmis, MW, Whiley DM, Thomas M, Mackay I, Williamson J. et al. (2004). A sensitive, specific, and cost-effective multiplex reverse transcriptase-PCR assay for the detection of seven common respiratory viruses in respiratory samples. J. Mol. Diagn. 6(2); 125-131. [ Links ]
10. Bellau-Pujol S, Vabret A, Legrand L, Dina J, Gouarin S. et al. (2005) . Development of three multiplex RT-PCR assays for the detection of 12 respiratory RNA viruses. J Virol Methods. 126(1-2); 53-63. [ Links ]
11. M'hadheb M, Harrabi M, Souii A, Jrad N, Gharbi J. (2015). Multiplex RT-PCR and Indirect Immnofluorescence Assay for Detection and Subtyping of human Influenza Virus in Tunisia. Curr Microbiol. (70); 324-329. [ Links ]
12. Fuller JA, Kariuki M, Bigogo G, Aura B, Maurice OO. et al. (2013). Association of the CT Values of Real-Time PCR of Viral Upper Respiratory Tract Infection With Clinical Severity, Kenya. Journal of Medical Virology. (Vol 85); 924-932. [ Links ]
13. Sidoti F, Bergalllo M, Costa C, Cavallo R. (2013). Alternative Molecular Test for Virological Diagnosis. Mol Biotechnol. (53); 352-362. [ Links ]
14. Pretorius M. Madhi S, Cohen C, Naidoo, Groome M. et al. (2012). Respiratory Viral Coinfections Identified by a 10-Plex Real-Time Reverse-Transcription Polymerase Chain Reaction Assay in Patients Hospitalized With Severe Acute Respiratory illness-South Africa, 2009-2010. Journal of Infectious Diseases (206); S159-S165. [ Links ]
15. Martins RB, Carney S, Goldemberg D, Bonine L, Cruz L. et al. (2014). Detection of respiratory viruses by real-time polymerase chain reaction in outpatients with acute respiratory infection. Mem. Inst. Oswaldo Cruz. Rio de Jainero. Vol (109) (6); 716-721. [ Links ]
16. Heim A, Ebnet C, Harste G, Pring P. (2003). Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol. 70(2); 228-239. [ Links ]
17. Gunson R, Collins T, Carman W. (2005). Real-time RT-PCR detection of 12 respiratory viral infections in four triplex reactions. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 33(4); 341-344. [ Links ]
18. Lu X, Chittaganpitch M, Olsen S, Mackay I, Sloots T. et al. (2006). Real-Time PCR Assays for Detection of Bocavirus in Human Specimens. J Clin Microbiol. 44(9); 3231-3235. [ Links ]
19. Boivin G, Cote S, Déry P, Serres G, Bergeron M. (2004). Multiplex Real-Time PCR Assay for Detection of Influenza and Human Respiratory Syncytial Viruses. Journal of clinical Microbiology. (Vol 42); 45-51. [ Links ]
20. Rheem I, ParkJ, Kim T, Wan J. (2012) Evaluation of a Multiplex Real-time PCR Assay for the Detection of Respiratory Viruses in Clinical Specimens. Ann Lab Med; 32; 399-406. [ Links ]
21. Dorak, T. (2006). Data analysis and reporting, in Real-Time PCR.Taylor & Francis; 39-61. [ Links ]
22. Fraga D., M.T., Fenster S. 2008. Real-time PCR, in Current protocols: essential laboratory techniques, W.E.A. Gallagher S. R. John Wiley & Sons, Inc., Hoboken, NJ; 10.3.1-10.3.34. [ Links ]