1. Introduction
Sugarcane is an important product for Colombian and world economy, representing 70% of the sugar total production. This plant is focus of study and research because of its different utilities in the food industry like the production of biofuels, liquors, fertilizers and fibers like paper. Sugarcane is susceptible to multiple microorganism’s attacks, including filamentous fungi such as Ceratocystis paradoxa, Colletotrichum falcatum, Fusarium verticillioides, Puccinia melanocephala, among others [1], several of this pathogens exist in Colombia invading the sugarcane crops, among these are the species from the genus Fusarium, most of them are highly pathogenic and toxic [2].
The particular fungus Fusarium verticillioides, acts rotting host plants infecting from the stem to the leaves during the first 4 to 6 months of life [1]; those plants who manage to survive this period are severely affected morphologically as well as physiologically. These symptoms reduce the productivity of crops, strongly and directly impacting the commercial production of sugarcane [3]. Also, reports of poisoning and development pathologies due to intake of products infected by F. verticillioides have been found [4] this related to the high production and release of mycotoxins generated by the Fusarium genus fungi, these mycotoxins are known as fumonisins [4]. The species Fusarium verticillioides is highlighted within the species of its genus for being a large producer of fumonisins, with the highest productivity. Fumonisins will not break down or denature in the presence of chemical agents neither at high temperatures, so it is almost certain that if these mycotoxins are present in sugarcane crops used for hu- man consumption products, they will reach up to processed products and finally they would be consumed, representing a serious risk to health. Due to the wide range of products that derives from sugarcane, the exposure of people to these toxins would be significantly high [5].
Fusarium verticillioides belongs to the Gibberella fujikuroi complex, this is a monophyletic taxon that includes at least 50 species from the genus Fusarium, with high similarity and overlapping morphological traits that makes their differentiation difficult and confusing [6]. It’s here where the importance of molecular identification lies, as relying only on morphological characterization may probably lead to errors on determining Fusarium species. This complex is also recognized because of the remarkable ability of its species to produce a wide range of mycotoxins and secondary metabolites that produce severe diseases in humans and animals [7], but the amount of toxins produced varies between species [8], therefore, knowing which taxonomic group was isolated in this work, it allows to establish the size of the threat that represents for food consumption security, crops safety and productivity.
To date in Colombia, Fusarium verticillioides has only been reported in the South of the country by [2], but there was not a formal report on the presence of the fungus Fusarium verticillioides as causal agent of Pokkah boeng disease and as a phytopathogenic agent for the case of sugarcane in Antioquia. For this reason, the principal aim of this project is to make a report of the presence of a sugarcane pathogenic fungus unknown in the region and contributing to molecular systematics and plant pathology studies.
2. Materials and methods
2.1 Isolation and cultivation of the fungus.
Sugarcane samples, which showed symptoms of red coloration and rotting characteristics typical of Pokkah Boeng disease, were collected in the municipality of Medellín (Antioquia, Colombia). Fungus-infested plant material was collected under the frame collection permit granted by the Agencia Nacional de Licencias Ambientales (ANLA) to CIBIOP research group (Universidad EAFIT, Resolution No. 1566 of 2014). The infested cane was acquired around the Poblado metro-station in Medellín, Colombia in a local sale of “guarapo” (sugarcane juice). The cane used in those beverages regularly comes from the municipalities of Barbosa and Copacabana where it is cultivated. The tissue samples from infected stems were immersed in 1.5% sodium hypochlorite for 1 minute, then in 75% ethanol for 30 seconds and finally washed in distilled water to eliminate the excess of these two compounds; all this with the aim of eliminating superficial pathogens as our microorganism of interest is a vascular fungus. Finally, disinfected tissue was cultured in acidified potato dextrose agar medium (PDA) (0,2% lactic acid) to prevent the growth of contaminant bacteria. A monoconidial culture was not obtained, nevertheless, what was observed through microscopy demonstrates the purity of the culture.
2.2 Sugarcane infection and control
The same infection methodology described in [9] was used. Three sugarcane plantlets (4 weeks old) were infected with the fungus strain EA-FP0013 and three others were used as control in greenhouse. Two horizontal cuts were made perpendicular from two vertical cuts separated by 5 mm each one of them at the upper part of the stem. Then a piece of a similar size was taken from the medium where the fungus was growing and put on the wound generated to favor the contact between the fungus and the plant. The controls were subjected to the same procedure but they were treated only with agar without any microorganism. This same test was also carried out using 8 banana plantlets (Musa acuminata cultivar Williams, 4 weeks old) in the same way, 4 as control and 4 infected, this to verify the possible host range of the EA-FP0013 strain. After 5 weeks of treatment tissue samples were taken and the same isolation process was made to determine if the same microorganism used for inoculation was obtained.
2.3 Molecular identiftcation
The isolate was grown in Sabouraud dextrose broth for one week at 28°C and constant agitation (200 rpm). Total DNA was extracted from the mycelium of the fungus following a CTAB protocol modified for the isolation of DNA from filamentous fungi, as described in [10]. The polymerase chain reaction (PCR) method was used to amplify marker genes ITS, Elon- gation Factor 1-α (EF) and β-Tubulin (Btub). The sequences of the primers used to amplify the ITS regions were taken from [11] and from [12],[13] for Btub and EF, see supplemental Table 1. The amplification programs were used with the following parameters: for ITS; 5 min at 95◦C, 35 cycles x (1 min 94 ◦ C, 1 min 55 ◦ C, 2 min 72 ◦ C), 5 min at 72 ◦ C. For B-tubulin 5 min at 95 ◦ C, 35 cycles x (35 sec 94 ◦ C, 55 sec 55.4 ◦ C, 2 min 72 C), 5 min at 72 ◦ C. For EF-1 it was 5 min at 95 ◦ C, 35 cycles x (1 min 94 ◦ C, 55 sec 50 ◦ C, 2 min 72 ◦ C), 5 min at 72 ◦ C. All reactions were performed in a C1000 Thermal Cycler (BioRad Technologies).
The amplification products were visualized on 1% agarose gel and quantified in Nanodrop (ThermoFisher Nano2000) for subsequent shipment to the Sanger sequencing service at the facility of the Universidad de los Andes, Colombia. The ab1 files obtained from the facility were analyzed (trimming bases below P20 and assembled to obtain consensus) using scripts in Python and tools from Biopython libraries [14]. The final consensuses from each gene were deposited in GenBank database of NCBI (National Center for Biotechnology Information - USA) under the following accession numbers: ITS (MH050788), Btub (MH899102), EF (MH899103).
2.4 Phylogenetic analysis
Two phylogenetic analysis were performed: 1) a single locus analysis based on ITS sequences and 2) a MultiLocus Sequence Analysis (MLSA) using three loci; the first corresponds to internal transcribed spacer region of ribosomal RNA locus (ITS-1, 5.8S and ITS-2). The other two correspond to conserved functional genes: β-Tubulin (B-tub) and Elongation Factor 1-α (EF). Sequences for comparison were retrieved using NCBI-BLAST against nucleotide database, using Biopython 1.67 modules [14]. The taxa used for each analysis an its accession numbers are listed in Supplemental File 1 presented as annexes.
The PhyPipe automated pipeline [15] (available at: https://gitlab.com/cibiop/phypipe/) was used for phylogenetic reconstruction by Maxi- mum Likelihood (ML) and Bayesian Inference (BI) methods. The pipeline of PhyPipe comprises alignment of DNA sequences with MAFFT 7.222, then partition analysis with PartitionFinder and phylogenetic reconstruction with RAxML 8.2.8, MrBayes 3.2.6 or Garli 2.01. For BI analysis, MrBayes was executed under the following parameters: two independent MCMC runs, four chains, 1,000,000 generations, 35% of the relative frequency of burning and sampling of 100. For ML analysis, Garli was executed by first performing a search ML (5 independent searches), 1000 bootstrap pseudoreplicates were made and mapped to the best ML topology using SumTrees from the DendroPy 4.1.0 package.
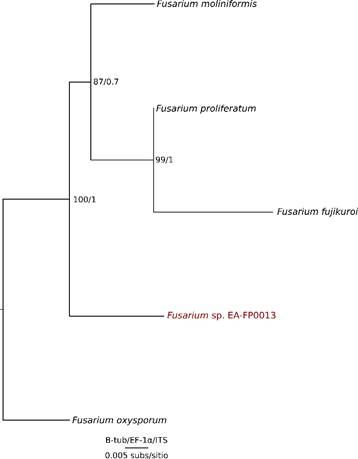
For the phylogenetic tree in Figure 4 all the other species belong to the Gibberella fujikuroi complex to have a balanced matrix. In addition the species used were only those that had the available sequences of the 3 genes (ITS, EF and B-tub) in the databases.
3.Results and discussion
3.1 Identiftcation and morphological characterization of isolated fungus
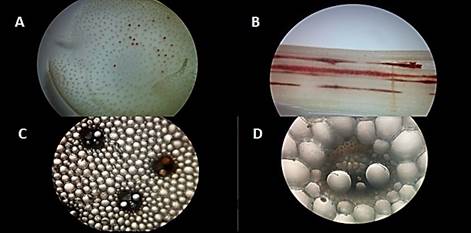
A filamentous fungus strain EA-FP0013 was successfully isolated in PDA medium from infected sugarcane tissue, whose characteristics coincide with the most representative characters of the strains of Fusarium verticillioides reported in the literature by [16],[17],[18],[19],[20]. The isolated fungus presents mycelial growth with colorations between pink, purple and red in the center with white edges [Figure 1A), radial growth, oval microconidia with flattened base and grouped in chains [Figure 1B and 1D). Also long, thin and septate macroconidia, with a curved apical cell [Figure 1C). The strain did not form chlamydospores at in vitro level in the evaluated medium.
F. verticillioides is one of the species of the Gibberella fujikuroi complex, so that, a differentiation of its morphological structures is needed in order to discard other fungus identities as Fusarium subglutinans, Fusarium sacchari and Fusarium pseudonygamai. According to previous articles, F. pseudonygamai forms large microconidia aseptate [17]; F. sacchari forms septate macroconidia thin, curved and with the apical cell curved in tip and basal cell in foot shape [20] and F. subglutinans forms curved, thin macro- conidia and almost cylindrical microconidia [21]. None of these characteristics were observed under the microscope in our isolated strain. Another important distinctive character observed in EA-FP0013 was the absence of microconidia grouped in chains in F. subglutinans, F. pseudonygamai and F. sacchari. All this evidence suggests that EA-FP0013 strain belongs to the species verticillioides, but as it is a complex of species, it important to corroborate using molecular identification and phylogenetic analysis.
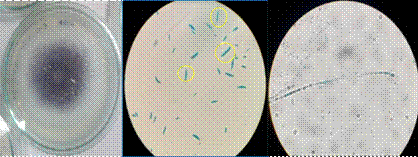
Figure 1 A. Fungus isolated from the sampled sugar cane, with purple and white mycelium. B. Stained tissue of the fungus visualized under the microscope (40X) septate macroconidia with curved apical cell in red and oval microconidia in yellow. C. Stained tissue of the fungus visualized under the microscope (40X) microconidia in chains.
3.2 Infection experiments
After obtaining a sample of infected cane and having isolated a fungus with the characteristics of a Fusarium strain, it was necessary to confirm if indeed the isolated fungus was the causative agent of the visible infection in the sugarcane sample. For this, Koch’s postulates were tested to determine its pathogenic activity. The results showed that isolated fungus EA-FP0013 was indeed the infectious agent generating the disease, causing the same symptoms of Pokka Boheng in the sugarcane used for this experiment. As can be seen in Figure 2, after the infection in the greenhouse, stems of healthy sugar cane developed the same red rot [Figure 2D and 2E) visible in Figure 2A and culturing of samples from those plants resulted in the isolation of a fungus [Figure 2B) with the same phenotypic characteristics as the one in Figure 1A. One control is visible in figure 2C, with no traces of the red rot. By looking tissue of the infected sugarcane under the stereoscope, it can be observed how the rot invades the stem fibers (Supplemental Figure 1A and 1B), and when observing the tissue under the microscope one can see how the fungus invades the phloem’s cells, this (Supplemental Figure, 1C and 1D) is the reason why the amount of juice that can be extracted from the sugarcane decreases when it is infected with Pokka boheng [21]. In all of the infected plants, the infection was extended through the vascular system to the leaves but it did not reach them and was neither found in the roots. This was due to the fact that the model of infection in the greenhouse does not allow verification in the foliar and radicular areas as this symptoms are usually visible in very advanced stages of the disease.
In addition to carrying out the infection of sugarcane plants in the greenhouse, banana plants were also infected under the same procedure, in order to get more information about the fungus’s host range. The result showed that banana plants did not develop the disease but presented a rapid hypersensitivity response after several weeks of being infected [Figure 2F), where a small part of the cellular tissue surrounding the site of the infection passed through cell death in order to restrict the growth of the pathogen. This also helped us discard the possibility of the strain being F. oxysporum as this species of Fusarium is widely spread and recognized as a banana pathogen.

Figure 2 A. Sample of sugar cane from which the fungus was isolated. B. Fungus isolated from the sample in picture E. C. Infection control plant from greenhouse experiment. D and E. Symptoms developed by plants weeks after infection in greenhouse. F. Banana plant weeks after infection in the greenhouse, it can be observed that the plant did not develop the disease.
3.3 Gene sequence analysis and molecular phylogenetics
An initial phylogenetic analysis was carried out with the ITS sequence from our fungal isolate and several other ITS sequences retrieved from GenBank, phylogenetic reconstructions were obtained from BI and ML analysis, obtaining in both cases the same topology [Figure 3]. This topology shows a polytomy among the Fusarium genus belonging to the Gibberella fujikuroi complex, but it does show phylogenetic difference with Fusarium proliferatum, this being different from the polytomic sequences (with high support: BV>75 PP>0.95). Sequences from the Gibberella fujikuroi complex also differentiate from F. oxysporum and F. subglutinans, which also form a well supported clade. The lack of resolution may be due to the state of conservation of the ITS genes within the genus, that is to say that in this locus sufficient mutations have not accumulated enough so that the species can be differentiated based sole on this gene marker [22]).
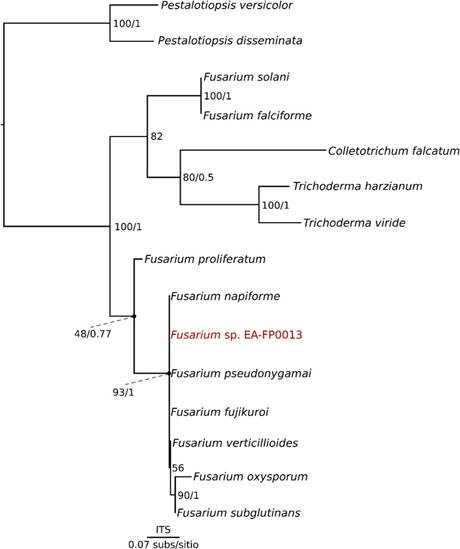
Figure 3 Phylogenetic tree derived from single analysis of related species of the genus Fusarium spp. using the nucleotide sequences of the ITS gene.
Looking for sequences of F. verticillioides in the databases, we only had ITS sequences in the GenBank, therefore F. moniliformis was used instead, but as the result shows, it does not group with the isolated sequence. It can be concluded that it is not F. fujikuroi, F. moniliforme, F. proliferatum, or F. oxysporum. In this point, there is no 100% certainty that the sequence isolated in this study is F. verticillioides, but there is sufficient evidence to say that it belongs to the Gibberella fujikuroi complex. Then, in order to give more resolution to this complex and differentiate EA-FP0013 from the other species, two other molecular markers were selected, EF and Btub. These markers have been used for many researchers around the world with successful results [23],[24],[25]. A phylogenetic analysis was carried out with concatenated genes and, through the ML and BI methodologies, a single phylogenetic tree was obtained [Figure 4]. With this resolution it was possible to differentiate the species of the Gibberella fujikuroi complex with good support levels.
Subsequently, the Btub and EF genes were used as molecular markers with the aim of solving the polytomy. As a result, the new topology shows us that the sequences belong to different species and our sequence of interest (Fusarium sp. EA-FP0013) is the sister group of the clade containing Fusarium moniliformis, which is the old denomination of Fusarium verticillioides [22],[26] [Figure 4], all of them belonging to the Gibberella fujikuroi species complex. It can also be observed that all the species mentioned above differentiate from Fusarium oxysporum, including our strain in particular, discarding the possibility that EA-FP0013 is F. oxysporum [27] [Figure 4], this allows us to define with greater certainty that the isolated sequence of the sugarcane belongs to the phytopathogenic Gibberella fujikuroi species complex, possibly to F. verticillioides species according the morphologic similarities between this species and our sugarcane iso- late [26]. Despite this, further studies are required in order to define the taxonomic status at species-level of this isolate. Finally, it was observed that F. oxysporum was not part of the complex, being this the reason why in the analysis that takes into account EF and B-tub genes this species was used as an outgroup. F. oxysporum is well reported as a pathogen of banana and other monocotyledons. Today this species counts with special attention thanks to owning races that devastate the cultivation of bananas. From this work we show that the isolated Fusarium strain does not belong to the F. oxysporum species, since both molecularly and pathologically it does not present the characteristics of this lineage
4. Conclusions
From a sample of sugar cane showing the symptoms of Pokkah boeng, a strain of phytopathogenic fungus belonging to the genus Fusarium and to the Gibberella fujikuroi complex, probably of the species Fusarium verticillioides, was isolated through morphological and molecular techniques. We validated the Koch’s postulates infesting cane plantlets and determined its pathogenic activity, concluding also that the EA-FP0013 strain was not pathogenic for banana plants. The presence of this pathogenic species was unknown, thus, this report opens the doors to different perspectives such as research into the biology of this pathogen and about the disease it produces, structuring mitigation and control strategies to avoid economic losses and food contamination problems, alert health authorities and Studies of the toxins produced by the fungus and its impact on the health of consumers, all of them of highly importance for the industry and public health.