INTRODUCTION
The Geminiviridae family is characterized by the presence of single-stranded, circular DNA genomes that are encapsidated in twinned isometric particles of about 18x30 nn. This family is into nine genera based on genome organization, vector transmission and host range: Curtovirus, Mastrevirus, Topocuvirus, Becurtovirus, Eragrovirus, Turncurtovirus, Capulavirus, Grablovirus and Begomovirus (Zerbini et al., 2017). The begomovirus genomes endemic to the Western Hemisphere (>322 species) consist of DNA-A and DNA-B components, each of 2.5-2.6 kb, are transmitted by members of a whitefly cryptic species complex [Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae)], and cause economically important diseases in most tropical and subtropical regions of the world (Brown et al., 2015; Gilbertson et al., 2016).
Many scientific reports suggest that weeds are potential sources of primary inoculum and play an important role as reservoirs and alternative hosts that spread viruses to new host crops and lead to major yield losses (Barreto et al., 2013; Prajapat et al., 2014; Vaca-Vaca et al., 2018). Therefore, the epidemiology of the geminivirus is incomplete without the study of weeds with crop-infecting begomoviruses that may have arisen from endemic weed-infecting viruses; there are known weed-infecting begomoviruses that can infect crops (Sanchez-Campos et al., 2000; Andrade et al., 2006). Wild hosts, such as weeds, are sites where begomoviruses carry out processes of genetic recombination or pseudo-recombination, decisive steps in the evolution of viruses. In addition to the processes of competition and selection, they can generate new variants of viruses with attenuated or higher pathogenicity in a short period of time (Padidam et al., 1999; Da Silva et al., 2011). Studies on viruses are of special importance to finding virus control measures in crops (Gallo et al., 2018).
Research on bipartite begomoviruses in Colombia is still very limited. Right now in Colombia, there are five reported virus species: bean dwarf mosaic virus (BDMV) (Gilbertson et al., 1991), bean leaf crumple virus (BLCrV) (Carvajal-Yepes et al., 2017), potato yellow mosaic virus (PYMV) (Martínez et al., 2008; Vaca-Vaca et al., 2012), and passionfruit leaf distortion virus (PDLV) (Vaca-Vaca et al., 2017), which have been isolated from beans, tomato and passion fruit crops, respectively. Recently, a new begomovirus was detected in chili peppers (Capsicum spp.) in Valle del Cauca but its identity is being studied (Vaca-Vaca et al., 2019b).
In addition, begomoviruses detection has been done in weeds collected from tomato crops in Valle del Cauca, such as Rivina humilis, Lapoerta aestuans, Lantana camara, Amaranthus dubius, Desmodium sp., Momordica charantia, Hybanthus attenuates, Verbena sp., Croton hirtus, Rhynchosia minima and Caesalpinia sp. (López-López et al., 2012; 2014). In 2018 and 2019, the complete genomes of begomoviruses were found in C. hirtus and R. minima, both new begomoviruses with the proposed names Croton golden mosaic virus (CroGMV) and Rhynchosia golden mosaic Colombia virus (RhGMCV), respectively. (Vaca-Vaca et al., 2018; López-López et al., 2019). Recently, new begomovirus hosts were identified in a study conducted in Valle del Cauca on weeds associated with chili pepper crops: Parthenium hysterophorus, Malvastrum sp., Sida acuta, Acalypha sp., Euphorbia hirta, R. minima and R. humilis (Vaca-Vaca et al., 2019a).
The increasing occurrence of begomoviruses in Colombia demands efforts to study their diversity in order to anticipate and monitor outbreaks, as well as to understand the evolutionary forces driving the emergence of novel begomoviruses in crop species previously reported as unaffected by these pathogens. Previously, a weed survey was conducted by the IPMA research group in tomato crops infected with PYMV in Valle del Cauca, where L. camara, A. dubius, R. humilis, Desmodium sp., R. minima, H. attenuatus, Verbena sp., C. hirtus and Caesalpinia sp. were identified as begomovirus hosts, but the identity of these viruses is still unknown to date (López-López et al., 2014). Therefore, the aim of this research was to characterize begomoviruses present in the weeds samples collected in 2014 along the edges of a tomato crop field located in four municipalities (Florida, Ginebra, Cerrito and Candelaria) in the Southeast of Valle del Cauca. The information obtained in this research could be useful for designing future effective strategies for the control of geminiviral diseases in crops.
MATERIALS AND METHODS
Sample collection: Weed samples were collected in tomato crops located in the southeast of Valle del Cauca (Colombia), and begomoviruses were detected with PCR (López-López et al., 2014). Eleven begomovirus positive weed samples were used in this research: four weed samples (leaves) were collected in the municipality of Florida: H. attenuates (AT18), Verbena sp. (AT19), C. hirtus (AT20) and L. camara (AT29); one sample was collected in the municipality of Ginebra: A. dubius (AT32); another five weeds were collected in the municipality of Cerrito: R. minima (two samples AT35 and AT36), R. humilis (AT38), Desmodium sp. (AT40) and Caesalpinia sp. (AT41); and the last sample was collected in the municipality of Candelaria: R. humilis (AT61).
Amplification and cloning of partial DNA-A component: Plant total DNA was extracted from leaf samples using the Dneasy Plant mini kit (Qiagen®). A 1.2 kb fragment was amplified with PCR with universal primers for members of the Begomovirus genus (Rojas et al., 1992). This fragment spans the 5´ region of AC1 (Rep) and coat protein (AR1) genes, and the entire intergenic region (IR) of the begomovirus. For each sample, a 1.2 kb fragment was cloned in pGEM T-Easy Vector (Promega®) and transformed in E. coli One Shot® (Invitrogen), according to standard procedures. Five clones of each fragment were completely sequenced at Macrogen Inc. (South Korea) with primer walking.
Sequence nucleotide analysis: Contigs of viral fragments were assembled using CLC Main Workbench, version 7.5 (Qiagen®), and were initially analyzed using the BLASTn algorithm (Zhang et al., 2000) to determine the viral species with greatest identity. The 1.2 kb fragments of the 5´ region of both the AC1 (Rep) and coat protein (Cp) genes and the intergenic region (IR) of all the clones were aligned using the MUSCLE algorithm in MEGA 7.0 (Kumar et al., 2016). To establish the identity of each sequence, the pairwise percentage nucleotide identity was calculated using Sequence Demarcation Tool Version 1.2 (SDTv1.2) (Muhire et al., 2014). Thirteen nucleotide sequences obtained in this study, together with the begomovirus sequences of the Abutilon and Squash clades, were aligned using the ClustalW algorithm with the default parameters. A phylogenetic tree was constructed with the neighbor-joining method with 2000 bootstrap replicates using MEGA 7.0. Phylogenetic trees were rooted with the sequence of a bipartite begomovirus, African cassava mosaic virus (ACMV, AF112352), from the Eastern Hemisphere (Nigeria) and a monopartite begomovirus, Tomato yellow leaf curl virus (TYLCV, X15656).
Confirmation of mixed viral infection with PCR: In order to confirm mixed viral infection in the samples of Desmodium sp. (AT40) and A. dubius (AT32), specific primers were designed using CLC Main Workbench, version 7.5 (Qiagen®), based on the nucleotide sequences PLDV (KT899302) and PYMV (JQ045706) deposited in the GenBank database. Following this strategy, the oligonucleotides PYMV-1: 5´-TTG-GGC-CAA-GAA-TAA-ACA-AGG-CCG-3´ / PYMV-2: 5´-ACC-ACC-TGT-GAC-CTT-GCC-ATA-GAA-3´; and MaraAF23-CP: 5´-TCC-GCT-CTC-TCT-TTC-TCT-TT -3´/ MaraAR405-CP: 5´-GGA-TAT-GTC-GTG-ACG-CTG-3´ were designed and using these pairs of primers, yield fragments of 500 bp and 467 bp of PYMV and PLDV, respectively.
RESULTS AND DISCUSSION
Six begomoviruses isolated from weeds in tomato crops were identified
From each weed, five clones were obtained and fully sequenced with primer walking. Each amplified clone carried a fragment of approximately 1.2 kb of DNA-A. The nucleotide sequences were assembled in CLC® Main Workbench v7 (Qiagen) and analyzed. Nine weeds presented five identical DNA fragments (100% between themselves), meaning each weed contained a unique virus, with the exception of A. dubius (AT32) and Desmodium sp. (AT40), which presented two different fragments, indicating two viruses in each weed. The analysis yielded a total of thirteen DNA fragments (Tab. 1). The analysis of these thirteen nucleotide sequences with SDTv1.2 generated a matrix of percentages of identity that showed the presence of six viral group clusters with a higher percentage of identity (> 94%) (Fig. 1).
Group (I) consisted of one virus isolated from C. hirtus (FR-20-Cr), which was different from other DNA fragments. Group (II) consisted of begomoviruses isolated from H. attenuatus (FR-18-Hyb) and R. humilis (FR-61-Riv), which shared a higher identity (99%). Group (III) had begomoviruses isolated from of L. camara (FR-29-Lan) and Desmodium sp. (FR-40.10-Des). Group (IV) contained one begomovirus isolated from Verbena sp. (FR-19-Ver). Group (V) corresponded to the isolate FR-32.1-Am, obtained from A. dubius. Finally, Group (VI) had the highest identity (99-100%) of the nucleotide sequences presented by the begomoviruses isolated from A. dubius (FR-32.6-Am), R. minima (FR-35-Rhy and FR-36-Rhy), R. humilis (FR-38-Riv), Desmodium sp. (FR-40.5-Des) and Caesalpinia sp. (FR-41-Cae).
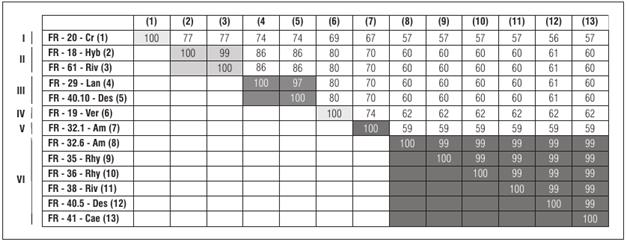
Figure 1 Identification of six viral groups with the highest percentage of identity (>94%) between the thirteen fragments isolated from weeds in this study. Percentages of nucleotide identity was calculated with SDTv1.2. For details and the nomenclature of each sample, see Tab. 1. The groups of begomoviruses are indicated with Roman numerals (I-IV)
The nucleotide sequences were analyzed using the BLASTn algorithm for sequence identity searches. The results of this analysis are shown in Tab. 1. DNA fragments isolated from H. attenuatus (FR-18-Hyb), C. hirtus (FR-20-Cr), and R. humilis (FR-61-Riv), which were collected in the municipalities of Florida and Candelaria, respectively, showed the highest percentage of identity (89%) for Bean chlorotic mosaic virus (BChMV, JN848772), a begomovirus isolated from bean in Venezuela (Fiallo-Olivé et al., 2013b). This result is remarkable since the FR-20-Cr fragment is composed of a separate identity group (group I) of the FR-18-Hyb and FR-61-Riv fragments (group II), which would indicate that both begomoviruses are related to BChMV but are different viral entities (Fig. 1). The FR-19-Ver fragment isolated from Verbena sp. showed the highest percentage of identity (89%) for the begomovirus datura leaf distortion virus (DaLDV-JN848773), isolated from the weed Datura stramonium in Venezuela (Fiallo-Olivé et al., 2013a).
Meanwhile, a DNA fragment (FR-29-Lan) isolated from L. camara and one (FR-40.10-Des) isolated from Desmodium sp. showed the highest value of identity (99%) for PDLV (KT899302), a begomovirus that affects passion fruit crops in Valle del Cauca (Vaca-Vaca et al., 2017). FR-32.1-Am, a DNA fragment isolated from A. dubius, showed the highest value of identity (99%) for PYMV (JN604019), a begomovirus isolated from tomato crops in Valle del Cauca (Martínez et al., 2008; Vaca-Vaca et al., 2012). These results showed for the first time the identification of three weeds that can be alternative hosts for begomoviruses that infect tomato and passion fruit crops in Valle del Cauca: L. camara collected in Florida and Desmodium sp. in Cerrito, both reservoirs for PLDV, while A. dubius, collected in Ginebra, is a host for PYMV. Similar results have been reported in Brazil, where Nicandra physaloides, Euphorbia heterophylla, Crotalaria spp. and Sida spp. weeds are natural reservoirs for the tomato severe rugose virus (ToSRV), a begomovirus that infects tomato crops (Barreto et al., 2013). Another example is the tomato yellow spot virus (ToYSV), a begomovirus that severely affects tomato crops at Brazil, which was identified in the alternative host Sida spp. (Ferro et al., 2017).
Table 1 Percentage of nucleotide identity for thirteen begomoviral fragments isolated from weeds in this study compared with begomoviruses deposited in the GenBank, calculated with the BLASTn program.
Finally, the DNA fragments belonging to group VI (FR-32.6-Am, FR-35-Rhy, FR-36-Rhy, FR-38-Riv, FR-40.5-Des and FR-41-Cae) presented a highest identity (89-90%) for Rhynchosia golden mosaic Yucatan virus (RhGMYV, EU021216), a begomovirus previously detected in R. minima in Yucatán, México (Hernández-Zepeda et al., 2010). It is remarkable that all these viral fragments were isolated from weeds collected in the same tomato crop located in the municipality of Cerrito. The nucleotide sequences of each viral fragment were deposited in the Genbank database (are described in Tab. 1).
In summary, six different begomoviruses were identified in thirteen DNA fragments obtained from eleven weeds collected in the municipalities of Florida, Ginebra, Cerrito and Candelaria (Tab. 1 and Fig. 1). Four of these viruses could be new begomoviruses not previously reported, based on the sequence identity for all other known begomoviruses to date and in accordance with the ICTV species demarcation criterion for the genus Begomovirus (≥91 % sequence identity) (Brown et al., 2015). In order to confirm this assumption, it is necessary to obtain the complete genome of each weed’s begomovirus (Tab. 1). This result confirmed the relevance of weeds as reservoirs for new unidentified geminiviruses that could become a limiting factor for economically important crops such as tomatoes. Further studies are required to explore the infectivity, geographical distribution and host range of new begomoviruses. Recently, our research group reported the complete genome sequence of begomoviruses isolated from C. hirtus and R. minima, confirming them as new begomoviruses (Vaca-Vaca et al., 2018; López-López et al., 2019).
The phylogenetic analysis showed that the thirteen DNA fragments were placed in two separated clades of begomoviruses: Squash leaf curl virus (clade Squash) and Abutilon mosaic virus (clade Abutilon) (Fig. 2).
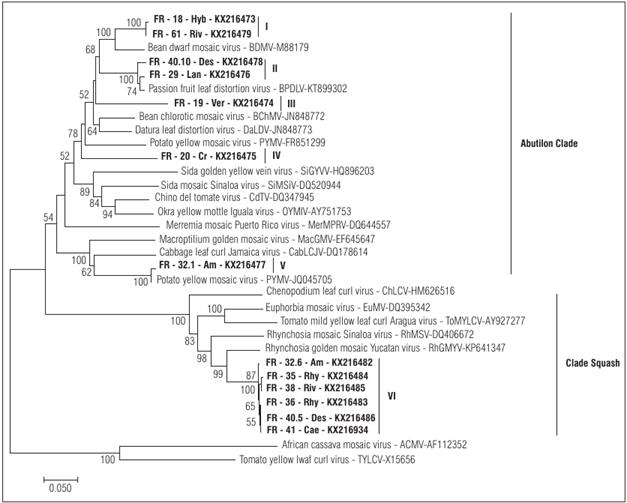
Figure 2 Phylogenetic relationships of thirteen begomoviruses isolated from weeds in this study with the most closely related begomoviruses of the Abutilon and Squash clades. Bold letters indicate the viral fragments isolated in this study (see Tab. 1 for details). The Roman numerals show the six viral groups identified in this study (see Fig. 1 for details). The bar below each tree indicates the nucleotide substitutions per site. The bootstrap consensus of the tree was inferred from 2000 replicates. Only bootstrap values above 50% are shown. ACMV (AF112352) and TYLCV (X15656) were used as an outgroup
The DNA fragments obtained from A. dubius (FR-32.6-Am), R. minima (FR-35-Rhy and FR-36-Rhy,), R. humilis (FR-38-Riv), Desmodium sp. (FR-40.5-Des) and Caesalpinia sp. (FR-41-Cae) were placed together with viruses belonging to the Squash clade. Meanwhile, the DNA fragments FR-18-Hyb, FR-19-Ver, FR-20-Cr, FR-29-Lan (PLDV), FR-32.1-Am (PYMV), FR-40.10-Des (PLDV) and FR-61-Riv were placed together with viruses belonging to the Abutilon Clade. In the particular case of DNA fragments, FR-18-Hyb and FR-61-Riv were most related to BDMV isolates, a begomovirus previously reported as affecting bean crops in Colombia (Gilbertson et al., 1991). The phylogenetic analysis showed that the DNA fragments FR-19-Ver and FR-20-Cr were placed on a monophyletic branch together with other begomoviruses and were closely related to isolates previously reported in tomato, bean and passion fruit crops in Colombia and Venezuela (Martínez et al., 2008; Vaca-Vaca et al., 2012; Fiallo-Olivé et al., 2013b; Vaca-Vaca et al., 2017). These results are in accordance with the pairwise sequence identity data (Tab. 1).
It is believed that the emergence of crop-infecting begomoviruses was the result of the horizontal transfer of indigenous viruses that infect weed hosts by the polyphagous B. tabaci biotype B vector. Geminiviruses exhibit high mutation rates and recombination frequencies, both within and between species, resulting in adaptative evolution. Recombination mechanisms are common among geminiviruses and contribute greatly to the evolutionary potential and local adaptation (Vaca-Vaca et al., 2012; Fargette et al., 2006; Padidam et al., 1999). For example, a number of natural begomovirus recombinants have been responsible for severe diseases and great economic losses in cassava in East Africa (Padidam et al., 1999). Another evolutionary mechanism that drives new geminivirus evolution and emergence is pse-udorecombination, which is generally restricted to the DNA genome components of closely related species. In some cases, recombinants and pseudo-recombinants have exhibited a new pathogenic phenotype that could be more virulent (Sanz et al., 2000; García-Andrés et al., 2006).
Mixed infection by begomoviruses in Desmodium sp. and A. dubius
The sequence analysis detected mixed infections in two weeds, Desmodium sp. and A. dubius (Tab. 1). Desmodium sp., collected in the municipality of Cerrito, had two begomoviruses, which were present in two different DNA fragments: one of which presented the highest identity for PLDV (99%), while the other one was more related to RhGMYV (89%). In the case of A. dubius, collected in the municipality of Ginebra, two different begomoviruses were present: one DNA fragment of these showed the highest identity with PYMV (99%), while the other DNA fragment one was related to RhGMYV (89%). The presence of PLDV and PYMV was corroborated by PCR using specific primers in Desmodium sp. and A. dubius. These specific primers amplified the coat protein gene (CP) for PDLV (500 bp) and PYMV (467 bp), respectively (Fig. 3). As a consequence, in Desmodium sp. and A. dubius, there were mixed infections by at least two begomoviruses. Mixed infections by different begomoviruses are common in non-cultivated host facilitating recombination events that may contribute to the frequent emergence of new species (Monde et al., 2010). Both viruses can interact in mixed infections, inducing more severe symptoms in crops, as reported by Jordá et al. (2007). Biologically, there are side benefits of mixed infections that can render some advances for begomoviruses, such as: a) increased rates of viral replication (Rentería-Canett et al., 2011); b) change or modification of tissue tropism (Alves-Junior et al., 2009); and c) efficient viral systemic movement and enhanced host begomoviral symptoms (Fondong et al., 2013).
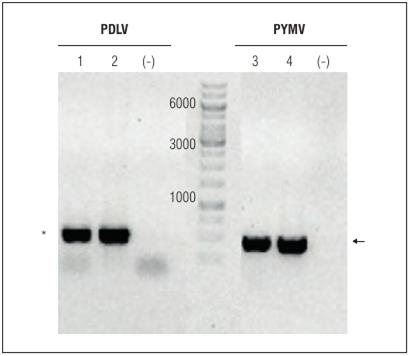
Figure 3 Detection of PLDV and PYMV with PCR in Desmodium sp. and A. dubius, respectively. Gel 1% agarose. M, molecular weight marker gene 1 kb DNA. 1. Desmodium sp. (sample AT40); 2. Passion fruit infected with PLDV (positive control); 3. A. dubius (sample AT32); 4. Tomato infected with PYMV (positive control). (-). Negative control. The asterisk indicates the expected 0.5 kb fragment for PLDV. The arrow indicates the expected 0.46 kb fragment for PYMV.
CONCLUSION
This study provided the partial molecular characterization of begomoviruses isolated from eleven weeds frequently found along the edges of tomato crop fields in four municipalities (Florida, Ginebra, Cerrito and Candelaria) in the southeast of Valle del Cauca. The results showed four probable new begomoviruses in H. attenuates, Verbena sp, C. hirtus, L. camara, A. dubius, R. minima, Desmodium sp., Caesalpinia sp. and R. humilis. These begomoviruses are more related to BDMV, BChMV, PLDV, DaLDV and RhGMYV, which were previously reported in crops and weeds in Colombia, Venezuela and Mexico.
PLDV and PYMV were detected in L. camara, Desmodium sp, and A. dubius, the first report of these weeds as alternative hosts for these viruses. Finally, there was evidence of mixed infections by two begomoviruses in A. dubius (PYMV/related RhGMYV) and Desmodium sp. (PDLV/related RhGMYV). Many studies have demonstrated that mixed infections by begomoviruses are the main source of novel recombinant geminiviruses that will constrain crop production in the future. As a consequence, the close monitoring and eradication of weeds such as L. camara, A. dubius and Desmodium sp. could reduce the incidence of begomoviruses in tomato and passion fruit crops. Further studies are required to explore the infectivity, geographical distribution and host range of this new, infecting begomovirus.















