INTRODUCTION
The cape gooseberry or golden berry (Physalis peruviana L.) is an herbaceous plant of the Solanaceae family (NRCS, 2020), with 1,311.8 ha planted in Colombia, mainly in the Boyaca, Cundinamarca and Nariño departments (Agronet, 2018), where it is grown commercially at altitudes between 1,800 and 2,800 m a.s.l., with medium temperatures between 13 and 16°C, 1,500-2,000 h year-1 of direct sunshine, and 1,000-1,800 mm year-1 of rainfall (Fischer and Melgarejo, 2020). Its fruit is a sugary berry preferred in countries such as Germany, Canada, Brazil, the Netherlands and Belgium, which are the main consumers (Hassanien et al., 2011). However, this crop is severely affected by vascular wilt, caused by Fusarium oxysporum, Schltdl., throughout Colombia (Galindo and Pardo, 2010; Enciso-Rodríguez et al., 2013; Rodríguez, 2013; Villarreal-Navarrete et al., 2017; Giraldo-Betancourt et al., 2020), generating losses of up to 100% (Smith, 2012; Mayorga-Cubillos et al., 2019) and forcing the movement of crops to other producing areas (González and Barrero, 2011).
Controlling this fungus is difficult because of the presence of inoculum in the soil, crop residues, infected plant debris and contaminated soils (McGovern, 2015). Plants can be infected by mycelium, conidia and chlamydospores (dormant propagules), which germinate when they come into contact with plant root exudates (Haglund and Kraft, 2001; Michielse and Rep, 2009; Zacky and Ting, 2013). In Colombia, the principal management strategy for this disease is treatment with flutriafol, propineb, iprodione and chlorothalonil, among other chemicals (ICA, 2020). However, their efficacy is less than 30% (Bennett et al., 2011). Therefore, these methods for controlling this pathogen and cultural practices have not been enough (McGovern, 2015).
The use of antagonistic microorganisms for biological control is a promising alternative that has been incorporated into disease management (Köhl et al., 2019).
Studies on tomatoes and bananas under greenhouse conditions have shown that non-pathogenic strains of F. oxysporum reduce the severity of this disease (Shishido et al., 2005). A similar effect has been reported with the addition of the mycorrhizal fungus Glomus intraradices (Akköprü and Demir, 2005). Cape gooseberry crops have used strains of Pseudomonas fluorescens for biocontrols, decreasing the incidence of this disease in plants (Urrea et al., 2011; Toloza-Moreno et al., 2020). Additionally, the biocontrol agents (BCAs) Brevibacillus brevis and Trichoderma harzianum have reduced the severity of the disease caused by F. oxysporum f. sp. lycopersici and F. oxysporum f. sp. cubense in tomatoes and bananas by 30 and 55%, respectively, and by 45% against F. oxysporum f. sp. phaseoli when was applied to bean seeds at greenhouse conditions (Carvalho et al., 2014; Bubici et al., 2019).
Another interesting approach that has been studied around the world for the control of Fusarium sp. uses suppressive soils, where the microbial community in the soil controls this pathogen and reduces the incidence of the disease. Siegel-Hertz et al. (2018) reported that Adhaeribacter, Massilia, Microvirga, Rhizobium, Rhizobacter, Arthrobacter, Amycolatopsis, Rubrobacter, Paenibacillus, Stenotrophomonas and Geobacter generated suppressive soils against F. oxysporum f. sp. lini in Linum usitatissimum. Likewise, it has been suggested that microorganisms isolated from the rhizosphere of a specific pathosystem provide better control of this disease in the same crop since they are already adapted to the host, as opposed to introducing organisms from other pathosystems (Lucy et al., 2004). The use of microorganisms in consortia from potentially suppressive soils reduced this disease more efficiently than the use of individual agents in tomato and radish crops (Bubici et al., 2019). The microbiome of soils can limit the presence of phytopathogens, so this study focused on selecting microorganisms that have been isolated from the cape gooseberry rhizosphere of potentially suppressive soils with a biocontrol potential against F. oxysporum f. sp. physali using a gnotobiotic system.
MATERIALS AND METHODS
Biological material
Commercial cape gooseberry (Physalis peruviana) Colombia ecotype seeds were used for the assays. The seedlings were disinfected by immersing them in 70% ethanol (2 min) and 3% sodium hypochlorite (20 min), followed by three washings with sterile distilled water. Afterwards, they were left to dry and germinate in a humid and dark chamber for 12 d at 25°C. During this disinfection procedure, no seeds showed symptoms of pathogen development.
The F. oxysporum f. sp. physali Map5 (Fox) strain was selected because of its high virulence in cape gooseberries (Rodríguez, 2010). Three small disks (0.5 cm) were taken from Potato Dextrose Agar cultures (PDA) that were incubated at 25°C for 8 d, inoculated in 300 mL of Potato Dextrose Broth (PDB), and grown at 25°C with constant agitation at 125 rpm for 7 d to obtain the microconidia. Subsequently, they were filtered through sterile muslin, and the inoculum was adjusted to a final concentration of 5·105 UFC/mLusing a Neubauer chamber.
Evaluation of the biocontrol activity of the microorganisms
The microorganisms used in this study were provided by the Microbial Culture Collection, maintained by Agrosavia. The isolates were originally obtained from rhizospheric soils from two cape gooseberry farms, one with organic management and the other with conventional management, located in the Gualmatan and Puerres municipalities, respectively, in the Department of Nariño (Colombia). These zones have a historical absence of Fusarium wilt and have been pointed out as possible sources for suppressive soils. The filamentous fungi were recovered in PDA (Potato Dextrose agar) at 25ºC, the bacteria were recovered with LB (Luria-Bertani) at 28°C for 24 h, and yeast was recovered on YM (Yeast Malt agar) at 25°C for 72 h.
The taxonomic identification of these isolates was done by sequencing with molecular marker genes. For the fungi and yeasts, DNA was extracted using a ZR Fungal/Bacterial DNA MiniPrep Kit™, and, for the bacteria, an InvitrogenTM PureLinkTM Genomic DNA Mini Kit was used according to the manufacturer’s instructions. Once the nucleic acids were obtained, the ITS region of the fungi and yeasts was amplified with a PCR using the primers R_ITS4_KY03 and ITS3_KY02 (Toju, 2012), and, for bacteria, the primers 515F and 806R were used to amplify variable region V3-V4 of ribosomal gene 16S rRNA (Caporaso et al., 2011). Subsequently, the products of this PCR were purified using magnetic beads (AgencourtAMPure XP beads) and were amplified with a second PCR with different combinations of barcode type primers for each sample to obtain a code for each one with simultaneous sequencing with pyrosequencing using the Illumina MiSeq® system.
For the 16S rRNA gene library preparation, the first PCR was performed in a final volume of 25 µL, containing 0.1 µL of Taq Platinum (Invitrogen), 0.75 µL of MgS04 50mM, 2.5 µL of Buffer-Mg 10X, 0.5 µL of dNTPs 10mM, 0.5 µL (10µM) of each forward and reverse primers with adaptor sequences, 2 µL of DNA and 18.15µL of Ultrapure Distilled Water (Invitrogen). The PCR was performed in a T100TM Thermal Cycler, BioRad, with the following conditions: initial denaturation at 94°C for 3 min, 35 cycles of denaturation at 94°C for 45 s, annealing at 50°C for 60 s and extension at 72°C for 90 s, with a final extension of 72°C for 10 min (Caporaso et al., 2011). Amplicons were visualized in agarose gels (1.5% p/v) in 1X TAE at 100 V, 30 min. All PCR products were purified following the protocol of Agentcourt® AMPure® XP.
In the second PCR, 5 µL of the previous amplicon were used as the template, with 1 µL (10µM) of each of the forward and reverse barcoding primer, 0.1 µL of Taq Platinum (Invitrogen), 0.75 µL of MgCl2 50mM, 2.5 Buffer-Mg 10X, 0.5 µL of dNTPs 10 mM and 14.15 µL of Ultrapure Distilled Water (Invitrogen) added. The amplification conditions were the same but with 12 cycles. The final PCR products were purified, and the quality was observed in agarose gels (1.5% p/v).
For the ITS gene library, the first PCR was performed in a final volume of 25 µL, containing 0.1 µL of Taq Platinum (Invitrogen), 1.5 µL of MgS04 50mM, 2.5 µL of Buffer-Mg 10X, 0.5 µL of dNTPs 10 mM, 0.5 µL (10 µM) of each forward and reverse primers with adaptor sequences, 2 µL of ADN and 17.4 µL of Ultrapure Distilled Water (Invitrogen). The following conditions were used: initial denaturation at 95°C for 2 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 60 s, with a final extension of 72°C for 5 min. The amplicons were visualized in agarose gels (1.5% p/v) in 1X TAE at 100 V, 30 min. The procedure for the second step PCR and product purification were as described above, adding 5 µL of the amplicon to the mixture and decreasing the cycles to 12 in the thermocycler program.
The purified DNA amplicons were quantified in a NanoDropTM 1000 Spectrophotometer, pooled and adjusted to the same concentration for the sequencing in the Illumina MiSeq System at the Microbial genomics laboratory of the Molecular Genetics and Antimicrobial Resistance Unit at the Universidad El Bosque (Bogota, Colombia).
The quality of the high throughput sequencing data was analyzed with Fastqc (Blankenberg et al., 2010) and compared to the GenBank database using the Blastn algorithm. Once the microorganisms were identified, a decision matrix was designed to eliminate those species that, according to scientific literature, had ecotoxicological or toxicological risks, keeping those that did not present any risks and that had been reported as having biocontrol activity.
Gnotobiotic system for the selection of potential biocontrol microorganisms
The biocontrol activity was evaluated following the methodology described by Moreno et al. (2014). Two 13 d-old cape gooseberry seedlings grown from disinfected seeds were placed in a plastic Petri dish (138.9 by 21.1 cm) with Gelified Nutrient Medium Agar - agar enriched with microelements (KI, H3BO3, MnSO4, ZnSO4, Na2MoO4 and CuSO4). The seedlings were placed 2 cm away from the periphery towards the center of the Petri dish (Fig. 1A).
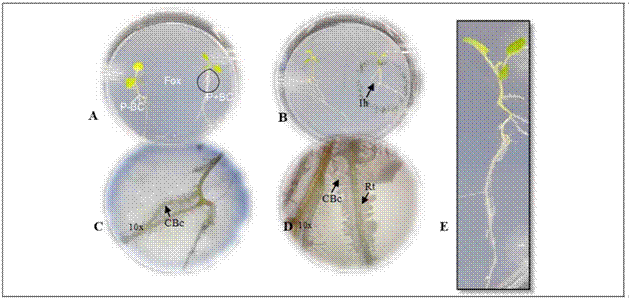
Figure 1. Gnotobiotic model and parameters evaluated in microorganisms with biocontrol potential. A. plant (P) - biocontrol (BC) interaction - Fox in the gnotobiotic model; P-BC: seedling without the biocontrol microorganisms, P + BC: seedling with the biocontrol microorganism, (Fox) Fusarium oxysporum. B. formation of inhibition halo of the pathogen (Ih). C. and D. colonization of the biocontrol microorganisms (CBc) in the radicle (Rt) with 10X magnifications. C., D. biofilm formation with 10X magnifications. E. plant growth.
The Biological Control Agent inoculum (BCA) was obtained with cell washing by scraping the surface of culture plates for the bacteria and yeasts (bacterial colonies grown on LB at 28°C for 24 h and yeast colonies on YM at 25°C for 72 h), adjusting the concentration to 1·108 UFC/mL and 1·107 UFC/mL, respectively. For the inoculum of the sporulating filamentous fungi, colonies grown in PDA at 25ºC for 8 d were scraped with a 0.1% Tween 80 solution, and the spore suspension was filtered through 2 layers of sterile muslin to remove mycelial fragments and adjusted to a final concentration of 1·106 UFC/mL. The inoculum concentration was determined according to previous reports (Bleve et al., 2006; Sahebani and Hadavi, 2008; Arrebola et al., 2010). The suspensions of non-sporulated fungi were obtained by fragmenting mycelium into an approximate size of 2 mm2 and scraping colonies with a 0.1% Tween 80 solution and diluting them 100-fold. A volume of 4 μL of each microorganism was applied to inoculate the seedlings on one plant, on the neck of the stem, which wasincubated at 25°C for 4 d (Fig. 1A). The center of the Petri dish was inoculated with 4 μL of a Fox suspension adjusted at 5·105 UFC/mL, and the dish was incubated at 25°C in a vertical position. Three repetitions (Petri dishes) were used for each microorganism with biocontrol potential. Two biological replicates were done.
Three different controls were considered in the experiment: the absolute control that consisted of seedlings without a biocontrol microorganism for Fox; the pathogen control with only seedlings inoculated with Fox; and the relative control consisting of seedlings treated with the biocontrol agent.
Ten days after incubation of the gnotobiotic dishes, the following parameters were evaluated to select microorganisms with biocontrol potential against Fox:(i) symptoms in the seedlings expressed as the presence of chlorotic and roller leaves, (ii) radicle colonization by Fox, (iii) Fox radial growth, (iv) formation of inhibition halo from biocontrol agent against Fox, (v) radicle colonization by fungi or bacteria biofilm formation and growth promotion expressed by total plant length (Fig. 1). A score was assigned to each of the parameters as shown in table 1.
Table 1 Parameters evaluated and the percent contribution to selection of microorganism with biocontrol potential in a gnotobiotic system.
| Features | Parameters | Score | Meaning | Contribution of the parameter to the feature (%)* |
|---|---|---|---|---|
| Protection against Fox | Symptoms development | 0 | Chlorotic and roller leaves | 0 |
| 1 | Extended leaves | 40 | ||
| Radicle colonization by Fox | 0 | Fox growth on the radicle | 0 | |
| 1 | Any growth of Fox on the radicle | 25 | ||
| Fox radial growth | 0 | Less growth of Fox in the presence of biocontrol agent than in its absence | 15 | |
| 1 | Greater growth of Fox in the presence of biocontrol agent than in its absence | 0 | ||
| Formation of inhibition halo from biocontrol agent against Fox | 0 | Not inhibition halo. Radicle colonization by Fox | 0 | |
| 1 | Halo produced by the biocontrol agent towards the pathogen. Not radicle colonization by Fox | 15 | ||
| Radicle colonization by fungi or biofilm formation by bacteria | 0 | Any growth of biocontrol agent on the radicle | 0 | |
| 1 | Growth of biocontrol agent on the radicle | 5 | ||
| Plant growth promotion | Total length plant | 0 | Less or same growth of plant length without biocontrol agent | 0 |
| 1 | Higher plant length growth in presence of biocontrol agent | 100 |
* Multicriteria analysis for the qualification both characteristics: protection against Fox and growth promotion according to the evaluated parameters.
In order to select biocontrol microorganisms, the evaluated parameters were compiled (Tab. 1, Fig. 1). A decision matrix analysis was used that weighed the different factors accordingly to the relative importance assigned to protection characteristics against Fox and plant growth promotion. This selection was based on the following criteria: review of toxicology risks and effects on non-target organisms, results of preliminary tests performed on mass production, results of their biocontrol activity against the pathogen and plant-growth promotion effect (Tab. 1).
Statistical analysis
A principal component analysis (PCA) was performed to determine the correlations between the data groups corresponding to microorganisms in growth promotion and protection against the pathogen, using R-Studio version 3.3 to detect differences and/or similarities between the microorganisms with potential biocontrol activity.
RESULTS AND DISCUSSION
Identification and selection of microorganisms with biocontrol potential
The AGROSAVIA National Microbial Culture Collection for Food and Agriculture provided 64 microorganisms isolated from the rhizosphere of potentially suppressive cape gooseberry soils in Nariño (Tab. 2), 29 were bacteria, and 35 were fungi. Nevertheless, 24 strains (15 bacteria and nine fungi) were discarded because they represented a risk for the environment (E) and for human and animal health (T) and/or they have not been reported as successful biological control agents (CB) (Tab. 2). As a result, 40 microorganisms, of which 14 were bacteria (five from a farm with conventional management and nine from an organic farm). i.e. 35%, and 26 were fungi (13 from each farm), i.e. 65%, which were evaluated in the gnotobiotic model against Fox (Tab. 2).
Table 2 Criteria for microorganism selection with biocontrol potential to be evaluated in a gnotobiotic model.
1 RT, microorganisms isolated from the rhizosphere of the farm with conventional management; RY, microorganisms isolated from the rhizosphere of the farm with organic management. E2(+),presence of ecotoxicological risk; E2(-),absence of ecotoxicological risk. T3(+),presence of toxicological risk; T3(-),absence of toxicological risk. CB4(+),successful reports of biocontrol activity; CB4(-),absence of unsuccessful reports of biocontrol activity. The selected microorganisms are highlighted in the table and are indicated with (+) and unselected with (-).
Evaluation of biocontrol activity of microorganisms
According to the principal component analysis (PCA), the plant growth promotion, expressed as total plant length (Tab. 1), presented 73.1% of the variability explained by the two characteristics total plant length and growth promotion (Fig. 2).
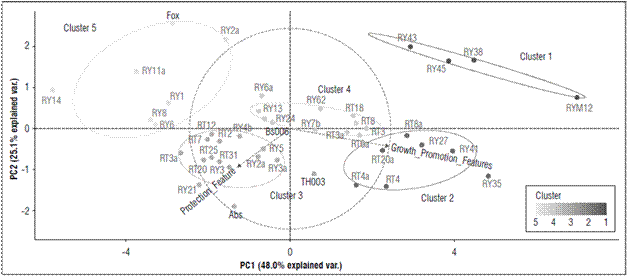
Figures 2. Principal component analysis (PCA) for the grouping of microorganisms with suppressive potential against F. oxysporum according to their similarity in the characteristics of biocontrol and growth promotion, obtained from the multicriteria table. The ellipses indicate the 95% confidence intervals for each cluster.
The microorganisms with biocontrol potential against Fox were grouped into five different clusters. Cluster 1 grouped 9.10% of the strains that, albeit with the lowest values for seedling protection (25%) in all evaluated parameters (formation of inhibition halo against Fox, symptom development, radicle colonization by fungi or biofilm formation by bacteria, radicle colonization by Fox and Fox radial growth), had the greatest effect on promoting growth in the seedlings with an average length of 11.2 cm (Fig. 2; Tab. 3). This cluster contained microorganisms (RY12-Paenibacillus peoriae, RY38-Serratia liquefaciens, RY45-Bacillus simplex, and RY43-Pseudomonas chlororaphis) that have strains that have been reported in many studies as plant growth promoting rhizobacteria (PGPR) (Kloepper et al., 1986; Raaijmakers and Weller, 1998; Pieterse et al., 2014; Bubici et al., 2019). Chin-A-Woeng et al. (2003) found that Pseudomonas spp. produces antifungal metabolites, such as phenazine, that favor growth promotion in Cucumissativus and Solanum lycopersicum plants. Likewise, other bacterial rhizosphere isolates from a potato crop, such as B. simplex, induced growth promotion in seedlings(Rete, 2011). S. liquefaciens has been reported as a plant growth-promoting rhizobacteria in cucumber (Kloepper et al., 1986), and P. peoriae has been reported as an antimicrobial agent that counteracts both fungi and phytopathogenic bacteria (Von der Weid et al., 2003). However, in this study, P. peoria had a growth promotion effect but no protection against Fox.
Table 3. Values of contribution to the selection of microorganism with and without a biocontrol potential agent in a gnotobiotic system.
| Code | Strain | Cluster | Protection against Fox + BC (%) | Growth promotion Fox + BC (%) | Growth promotion Fox + BC (cm) |
|---|---|---|---|---|---|
| RY12 | Paenibacillus peoriae | 1 | 25 | 100 | 9.4 |
| RY38 | Serratia liquefaciens | 1 | 25 | 100 | 9.2 |
| RY45 | Bacillus simplex | 1 | 25 | 100 | 13.0 |
| RY43 | Pseudomonas chlororaphis | 1 | 25 | 100 | 13.0 |
| RY35 | Lysinibacillus sp. | 2 | 25 | 100 | 10.6 |
| RY41 | Bacillus simplex | 2 | 25 | 100 | 12.4 |
| RY27 | Bacillus foraminis | 2 | 25 | 100 | 11.6 |
| RT8a | Rahnella sp. | 2 | 25 | 100 | 11.3 |
| RT20a | Bacillus thuringiensis | 2 | 25 | 100 | 13.1 |
| RT4a | Trichoderma sp. | 2 | 25 | 100 | 11.9 |
| RT4 | Trichoderma gamsii | 2 | 25 | 100 | 14.6 |
| Abs | Absolulo | 3 | 75 | 100 | 9.6 |
| TH003 | Trichoderma koningiopsis | 3 | 60 | 100 | 8.2 |
| RY21 | Bacillus subtilis | 3 | 60 | 100 | 10.9 |
| RY3a | Scopulariopsis brevicaulis | 3 | 60 | 100 | 10.1 |
| RY5 | Trichoderma viride | 3 | 60 | 100 | 8.8 |
| RY2a | Trichoderma gamsii | 3 | 60 | 100 | 8.4 |
| RY4b | Trichoderma gamsii | 3 | 60 | 100 | 8.2 |
| RY3 | Beauveria bassiana | 3 | 50 | 100 | 9.3 |
| RT31 | Bacillus sp. | 3 | 55 | 100 | 9.7 |
| RT3a | Mucor racemosus | 3 | 55 | 100 | 8.8 |
| RT20 | Chaetomiaceae sp. | 3 | 55 | 100 | 9.0 |
| RT25 | Chaetomiaceae sp. | 3 | 55 | 100 | 9.1 |
| RT2 | Penicillium cremeogriseum | 3 | 55 | 100 | 8.3 |
| RT7 | Penicillium janthinellum | 3 | 55 | 100 | 9.7 |
| RT12 | Purpureocillium lilacinum | 3 | 75 | 100 | 10.4 |
| Bs006 | Bacillus subtilis | 4 | 60 | 100 | 9.3 |
| RY25 | Bacillus subtilis | 4 | 60 | 100 | 12.3 |
| RY13 | Rhodococcus sp. | 4 | 60 | 100 | 9.2 |
| RY6a | Podospora setosa | 4 | 40 | 100 | 9.0 |
| RY62 | Debaryomyces vindobonensis | 4 | 40 | 100 | 9.6 |
| RY7b | Plectosphaerella plurivora | 4 | 60 | 100 | 8.7 |
| RT3 | Acinetobacter rhizosphaerae | 4 | 45 | 100 | 11.5 |
| RT6 | Bacillus subtilis | 4 | 60 | 100 | 9.0 |
| RT3 | Trichoderma koningiopsis | 4 | 60 | 100 | 9.6 |
| RT8 | Trichoderma koningiopsis | 4 | 75 | 100 | 11.2 |
| RT18 | Umbelopsis sp. | 4 | 45 | 100 | 11.0 |
| Fox | Fusarium oxysporum | 5 | 25 | 0 | 8.2 |
| RY2 | Trichoderma sp. | 5 | 25 | 0 | 6.4 |
| RY1 | Trichoderma koningiopsis | 5 | 25 | 100 | 8.3 |
| RY6 | Trichoderma ghanense | 5 | 25 | 0 | 7.9 |
| RY8 | Trichosporon lignicola | 5 | 25 | 0 | 6.5 |
| RT11a | Humicola grisea | 5 | 5 | 0 | 7.6 |
| RT14 | Doratomyces asperulus | 5 | 25 | 0 | 5.5 |
* The contribution of protection characteristics against Fox and promotion of growth in seedlings without biocontrol was zero because of the absence of the microorganism.
Cluster 2 grouped 15.9% of the microorganisms that generated a less protective effect on the seedlings (score of 25%) than the positive effect in terms of growth promotion (an average length of 12.2 cm) (Tab. 1; Fig. 2; Tab. 3). This cluster contained the microorganisms RY35-Lysinibacillus sp., RY41-Bacillus simplex, RY27-Bacillus foraminis, RT8a-Rahnella sp., RT20a-Bacillus thuringiensis, RT4a-Trichoderma sp. and RT4-Trichoderma gamsii. Previous studies have reported the presence of B. simplex and R. aquatilis in the rhizosphere of Vitis spp., which facilitate nitrogen fixation and promote plant growth in Solanum tuberosum and Vitis spp. (Chen et al., 2007; Velivelli et al., 2015). Singh et al. (2015) reported that B. foraminis, not only exerts a protective effect against Fusarium sp., but also favors the promotion of growth in Zea mays. Additionally, L. fusiformis, Lysinibacillus spp. and B. thuringiensis have been described as potential biocontrol agents of F. oxysporum f. sp. ciceri race 1 in Cicer arietinum (Singh et al., 2014). Similar results were found in the present study. Moreover, this study confirmed the biocontrol activity of Trichoderma sp. since T. gamsii and other species from this genus had a control effect against Fox in the cape gooseberry seedlings (Fig. 1B).
Cluster 3 included 34.1% of the microorganisms with high values in terms of seedling protection, a high score of 50% in these parameters, and a positive effect in terms of growth promotion (average length of 9.2 cm), but there were no differences between plant length (Tab. 1; Fig. 2; Tab. 3). This cluster contained microorganisms with biocontrol potential, such as TH003-Trichoderma koningiopsis, RY21-Bacillus subtilis, RY3-Scopulariopsis brevicaulis, RY5-Trichoderma viride, RY2-T. gamsii, RY4b-Trichoderma gamsii, RY3-Beauveria bassiana, RT31-Bacillus sp., RT3a-Mucor racemosus, RT20-Humicola grisea, RT25-H. grisea, RT2-Penicillium cremeogriseum, RT7-Penicillium janthinellum and RT12-Purpureocillium lilacinum. Some species of the Trichoderma genus are effective biocontrol agents (Schirmböck et al., 1994; Lorito et al., 1996; Howell, 2003), and T. koningiopsis, T. viride, T. gamsii and S. brevicaulis have been reported as antagonistic agents of Fusarium culmorum in Zea mays (Luongo et al., 2005). The Th003 strain of T. koningiopsis is the active ingredient of Tricotec®, a product registered in Colombia by Agrosavia for the control of F. oxysporum f. sp. lycopersici in tomato crops (Cotes et al., 2018). On the other hand, B. bassiana, M. racemosus and Penicillium spp. have been reported as mycoparasites on F. oxysporum f. sp. radicis-lycopersici (Palma-Guerrero et al., 2008). H. grisea and Purpureocillium sp. have been reported as biocontrol agents because of the production of antibiotics (Hubbard et al., 1982; Di Pietro et al., 1992) that inhibit pathogens such as F. culmorum and Pythium ultimum (Galindo-Flores et al., 2005). Asaka and Shoda (2002) found that B. subtilis produces antibiotics, such as iturin A and surfactin, that counteract pathogens such as Rhizoctonia solani in tomato plants.
Cluster 4 concentrated 25% of the microorganisms that generated the highest protection values against Fox (Score highest than 60%) and showed a positive effect in growth promotion (average length of 10 cm) (Tab. 1; Fig. 2; Tab. 3). The microorganisms in this cluster were Bs006-Bacillus velezensis, RY25-Bacillus subtilis, RY13-Rhodococcus sp., RY6-Podospora setosa, RY62-Debaryomyces vindobonensis, RY7b-Plectosphaerella plurivora, RT3-Acinetobacter rhizosphaerae, RT6-Bacillus subtilis, RT3-Trichoderma koningiopsis, RT8-Trichoderma koningiopsis, and RT18-Umbelopsis sp. Previous studies have shown that Bs006, a strain of B. velezensis, is an excellent Fox inhibiting microorganism because of antibiotic secretions (Moreno et al., 2014) and that Rhodococcus sp. inhibits Pythium aphanidermatum under in vitro conditions because the enzymes are capable of degrading the cell wall (El-Tarabily, 2006), which could explain the control activity of this microorganism against Fox. In contrast, Debaryomyces sp., Plectosphaerella sp., Umbelopsis sp. and Acinetobacter sp. have been widely studied as growth promoters (Rokhbakhsh-Zamin et al., 2011; Zhao et al., 2014), and Podospora sp. has been reported as an abundant microorganism in healthy Musa sp. and Pyrus sp. soils and has been associated with the suppression of vascular wilt caused by Fox (Xu et al., 2012; Huang et al., 2017).
Cluster 5 represented 15.90% of the microorganisms, which did not generate a protection effect against the pathogen in the seedlings or an effect on growth promotion (average length of 7.2 cm) (Fig. 2; Tab. 3). This group had the treatment that only contained Fox (pathogenic control). This cluster contained the microorganisms RY2-Trichoderma sp., RY1-Trichoderma koningiopsis, RY6-Trichoderma ghanense, RY8-Trichosporon lignicola, RT11-H. grisea and RT14-Doratomyces asperulus. In contrast to cluster 4, the two strains of Trichoderma from this group did not have a protective effect against the pathogen. Di Pietro et al. (1992) reported that H. grisea and D. asperulus are biocontrol agents because of the production of antimicrobial substances that counteract F. culmorum. Nevertheless, the strains in this cluster did not protect against the pathogen. On the contrary, some of them favored Fox because they allowed abundant growth and development of vascular wilt symptoms.
CONCLUSION
The biocontrol activity and growth responses of the cape gooseberries to the treatments with the evaluated rhizospheric microorganisms varied with the isolate-type and the origin of the isolates. The majority of the bacteria obtained from the organic farm expressed a biocontrol activity and positively affected plant growth, whereas the fungal isolates with a biocontrol activity and plant growth promotion were similar for the two farms. This effect reflects the capability of different microorganism species to counteract this pathogen and promote plant growth. However, a practical application of these results should be further evaluated in field experiments. Based on our results, Bs006-Bacillus velezensis, RY25-Bacillus subtilis, RY13-Rhodococcus sp., RY6-Podospora setosa, RY62-Debaryomyces vindobonensis, RY7b-Plectosphaerella plurivora, RT3-Acinetobacter rhizosphaerae, RT6-Bacillus subtilis, RT3-Trichoderma koningiopsis, RT8-Trichoderma koningiopsis, are RT18-Umbelopsis sp. are promising F. oxysporum control treatments for cape gooseberries (more than 60%), which should be evaluated both individually and in consortia under greenhouse conditions.















