INTRODUCTION
Soil constitutes a very complex system where physical, chemical and biological processes take place in which microorganisms carry out important activities for the ecosystemic maintenance (Urgiles-Gómez et al., 2021). Currently, the implementation of intensive agriculture has led to the development of diseases, nutrient deficits, soil degradation, loss of biodiversity, ero sion, among others (Pandey et al.,2019; Posada et al.,2021). One of the strategies to counteract this problem lies in the use of microorganisms with agricultural potential and among the most promising are plant growth promoting rhizobacteria (PGPR) and arbuscular mycorrhizal fungi (AMF) (Urgiles-Gómez et al.,2021); these microorganisms have the ability to minimize depen dence on chemical fertilizers and pesticides, thus optimizing soil fertility and quality (Zuluaga et al.,2020; Issifu et al., 2022), are also attributed with the ability to offer resistance to pathogens and environmental stress (Tabacchioni et al., 2021) and facilitate the stimulation of plant growth through mechanisms such as biological nitrogen fixation (BNF), phytohormone production, syn thesis of antibiotics and organic acids related to plant growth promotion, phosphate solubiliza tion, siderophore production, cellulase and protease synthesis, among others (dos San tos et al., 2022; Mghazli et al., 2022; Sachman-Ruíz et al., 2022).
Thus, the soil contains microbial populations including bacteria, which play an important role in the cycling of organic matter and nutrient availability, and also participate in ecological processes that regulate plant growth (Tang et al.,2023), because soil microorganisms play a fundamental role in the maintenance and regulation of the biological cycles of soil elements and there fore in crop yields (Fan et al.,2021). In addition, the microorganisms that inhabit the soil have an incidence in the formation and stabilization of aggregates, due to this, this type of organisms are related to the processes of soil construction (Cavael et al., 2020). Soil bacteria contribute to the decomposition of plant and animal residues to form compost, which improves soil structure, lead ing to the retention of nutrients and moisture, considered important factors in sustainable agricul ture (Tanya and Leiva-Mora, 2019). The activity of enzymes produced by bacteria is linked to the physicochemical properties, diversity and microbial composition of the soil, which means that they act as important indicators of soil productivity and biological activity (Alkorta et al.,2003). It has even been claimed that indigenous soil bacteria can reduce environmental pollution by pro ducing and utilizing enzymes that metabolize the pollutant, using it as a source of nutrients, whether organic or inorganic (Munawar et al.,2023).
It is important to make use of potential microbial populations, as they increase the restoration and development of soil components, maintaining the balance of biological interactions that favor agricultural production, applied as a natural and sustainable way with the environment (Gutiérrez-Calvo et al., 2022). In this sense, the consequences generated by the excessive use of agro chemicals are reduced, increasing the biological diversity of the soil (Aldonate et al., 2019; Diaz et al.,2019). In addition, the mechanisms used by PGPRs are useful for the productivity of differ ent crop species in the face of abiotic stresses such as salinity and drought (Jatan et al., 2019; Vu rukonda, 2020; Bécquer, 2022). Thus, the identification of soil microbial populations is of great value, since they can be used as biofertilizers and biocontrollers, depending on their metabolism and functionality within the strategy of sustainable agricultural production (Cruz et al., 2021). One way to identify microbial groups that are potentially beneficial in agriculture is by molecular techniques, which favor knowledge of the genes involved in obtaining productive systems fo cused on sustainable agriculture (Díaz-Rodríguez et al., 2021; Orozco-Mosqueda and Santoyo, 2021).
Molecular identification of microorganisms with agricultural potential is a topic of great significance, since it allows to make known to all those involved in agricultural production the dif ferent molecular tools that can be used for the identification of promising microorganisms with potential for agriculture. To this purpose, it is necessary to group these techniques and carry out a bibliographic and bibliometric mapping, which facilitates the development of the subject matter based on scientific documents, countries, authors, and journals that stand out in the area of study. It is important to mention that the Scopus and Web of Science databases were reviewed and no review articles were found that specifically compile the most used molecular biology techniques for the identification of PGPR and bacteria with agricultural potential, however in Scopus only one review article was reported, which was carried out by Urgiles-Gómez et al. (2021), who compiled research on the isolation and identification of PGPR where, in addition to using conventional methodologies, molecular analysis was performed, determining the presence of Azospirillum, Pseudomonas, Bacillus and other microorganisms in the rhizosphere of coffee plants (Coffea spp.), which are considered potential biofertilizers.
Thus, the present study aimed to develop a systematic and bibliometric mapping of the research carried out, in which molecular techniques are applied in soil microbiology, for the identification of bacteria with agricultural potential (MTIBAP). For the development of the objective, a bibliographic search was carried out through the Web of Science (WoS) and Scopus databases. For this purpose, it was filtered in the years 2000-2022 and the information resulting from the search was processed by means of the R studio software. Subsequently, a bibliometric mapping was carried out by means of a network study, listing the most relevant documents; to achieve this, the metaphor of the tree of science was used, which is made up of three parts: root, trunk and leaves. In this way, the main lines of research of this study were determined. Finally, the most significant studies on each topic were investigated. This bibliometric document was developed in four important phases: 1) Studies were selected, analyzed and verified through the use of search criteria determined for the purpose of this research. 2) A beginning evaluation and analysis of the theory was developed. 3) The bibliometric study was carried out and the relationships. 4) Deter mining words, limitations and suggestions for further research related to the topic of MTIBAP were noted.
MATERIALS AND METHODS
The strategy proposed in this study consisted of two stages: first, a bibliometric search was carried out in the Scopus and Web of Science (WoS) databases, which revealed scientific research focused on soil microbiology. Secondly, research related to molecular techniques used for the identification of microorganisms with agricultural potential was recognized. Thus, five biblio metric parameters were used to carry out these searches: citation analysis, co-occurrence of words, co-citations, co-authors and bibliographic coupling (Zupic and Čater, 2015). It is necessary to mention that Scopus and Web of Science are bibliographic databases that allow us to ob tain the type of information necessary to carry out this review (Echchakoui, 2020), as well as pro viding information at the international level (Zhu and Liu, 2020; Pranckutė, 2021). The search criteria are listed in table 1.
Table 1. Search criteria.
| Databases | Web of Science (WoS) | Scopus |
|---|---|---|
| Consulting period | 2000 - 2022 | |
| Consulting date | 17/01/2023 | |
| Search criteria | Title | |
| Journal type | Alls | |
| Search terms | (“Plant-Growth-Promoting Rhizobacteria” or “Plant growth-promoting bacterial endophytes” or “Plant growth promotion by phosphate solubilizing bacteria” or “Plant growth-promoting” or “plant growth-promoting bac terium” or “plant growth promoting fungus” or “plant growth promoting fungi” or “PGPF”) and (“molecular identification” or “hybridization” or “sequencing” or “PCR” or “polymerase chain reaction” or “real-time PCR” or “qPCR” or “Reverse transcriptase polymerase chain reaction” or “RT- PCR” or “Digital PCR” or “Microarrays” or “Genome sequencing” or “Sanger sequencing” or “Massively parallel sequencing” or “SMP” or “Next-generation sequencing” or “NGS” or “DGGE” or “Pyrosequencing” or “DNA probe hybridization” or “Denaturing gradient gel electrophoresis” or “Maldi tof” or “RFLP” or “PCR-RFLP” or “RAPD” or “loop-mediated isothermal amplification” or “LAMP” or “restriction fragment length poly morphism fragment* length polymorphism” or “identification” or “molecu lar identification” or “isolation” or “characterization” or “selection”) | |
| Results | 373 | 479 |
| Total results | 527 | |
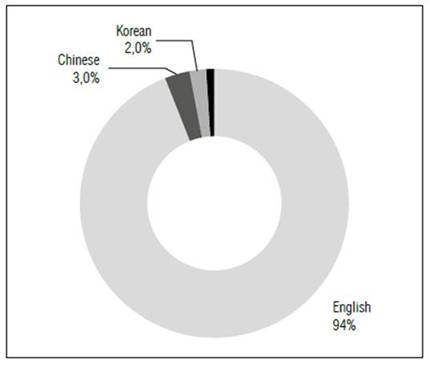
After using the R studio software, the results obtained are 479 articles in Scopus and 373 in Web of Science, which after joining and discarding duplicate articles, gave a total of 527 records, which means between the two databases an overlap of 61.66%, showing the relevance of applying them together. As a result, it was found that 94% of the research published on this subject re lated to Web of Science and Scopus is in English and 3, 2, 1% and less than 1% are in Chinese, Korean, Portuguese and others, respectively (Fig. 1). The argument is because both journals and writers publish their studies in English to increase the H-index of these databases (Vera-Baceta et al., 2019). The software used for the bibliographic analysis is Bibliometrix (Aria and Cuccurullo, 2017), this is a free access application, which allows working with different databases (Scopus and Web of Science), its utilities are diverse, and it has been applied and evidenced by other stud ies (Tani et al.,2018; Landinez et al., 2019; Duque-Hurtado et al.,2020; Acevedo et al.,2021; Di Vaio et al., 2021; Duque et al., 2021c; Queiroz and Wamba, 2021; Secinaro et al., 2022; Ra belo-Flórez, 2023).
Network analysis
By means of the R study software, replicated documents from the Scopus and Web of Science databases were excluded. Then, once the selected documents were obtained, the bibliographic sources were used in a grid of citations to build the graph theory, which consists of schematically representing the relationship between each of the appropriate documents that are part of this review (Wallis, 2007; Yang et al., 2017).
Three bibliometric indicators were taken into account, consisting of: the indegree, which refers to the greatest number of citations made by other authors about the article; the outdegree, which indicates the number of citations in which a given group uses ideas from other authors (Wallis, 2007), Therefore, they are not cited since they are recent articles; and finally, the betweenness that points out both the citation made by the authors of the article and the citation made in other documents about these same articles (Freeman, 1977; Zhang and Luo, 2017).
As a result, it was possible to acquire a mesh of data on the research on the subject of this study, acquired through Scopus and Web of Science. For this purpose, the Gephi software was used (Bastian et al., 2009), which facilitated to obtain in a representative way the map of cocitations, which confers to analyze the representation or the position that the researches on the MTIBAP topic acquire within the scientific studies, facilitating the visual obtaining of dynamic networks (Bastian et al.,2009; Gurzki and Woisetschläger, 2017; Zuschke, 2020).
The metrics of the bibliometric indicators were quantified for each of the corresponding network records, with the purpose of ordering the papers using the metaphor of the tree of science (Robledo et al.,2014; Valencia-Hernández et al.,2020). By means of this representation, three classifications can be obtained: the first one, the bases or roots (indegree), which integrates the most important studies or research within the subject of the study in question, which are highly referenced, but which do not cite others (Wallis, 2007); the second is the trunk (betweenness), where the articles or documents that are cited and also taken as a reference by others are located (Zhang and Luo, 2017). This section compiles the studies that relate the theoretical part of the bases or roots with the emerging research in the area of study and, thirdly, the out-of-degree studies, which correspond to current studies that cite the others but are not referenced (Wallis, 2007) and that also catalog current inclinations in the studied area. Thus, the methodological development of this study has been carried out and ratified by other authors in previous studies (Duque and Cervantes-Cervantes, 2019; Duque-Hurtado et al.,2020; Buitrago et al., 2020; Clavijo-Tapia et al., 2021; Duque et al., 2021a; Duque et al., 2021b; Ramos-Enríquez et al., 2021; Torres et al., 2021; Trejos-Salazar et al., 2021; Barrera et al., 2022; Rabelo-Flórez, 2023).
RESULTS
Bibliometric analysis
The bibliometric production analysis and evaluation were carried out using the bibliometric procedures described by Zupic and Čater (2015), with modifications. The Web of Science (WoS) and Scopus databases were used simultaneously, due to the fact that they provide more extensive access to the bibliographic content (Echchakoui, 2020; Pranckutė, 2021), as well, that these are the main international databases (Bar-Ilan, 2010; Zhu and Liu, 2020), and the relevance of scientific production related to bacteria of agricultural potential that is published in therein.
Historical analysis of scientific production
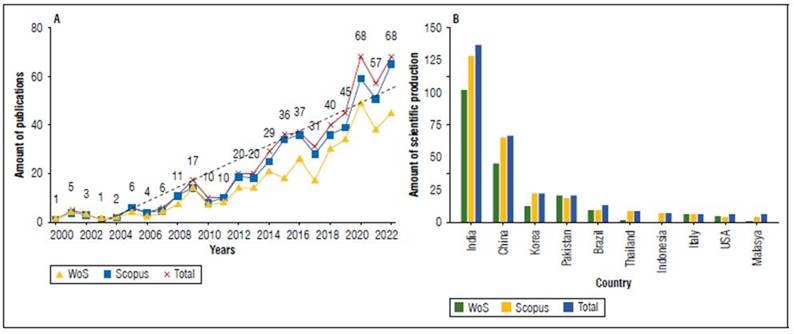
Figure 2A shows the number of scientific papers that have been published in relation to MTIBAP in the years 2000-2022. The results showed a total of 527 scientific documents through the application of the Web of Science and Scopus databases; in addition, it is possible to show that the subject under study indicates an upward trend, consistent with the significant increase in the number of articles published, so it is feasible to determine that research in this area of study is booming, due to the fact that in the last five years 52.75% of the scientific production has been published and an annual growth rate of 17.4%. Thus, this background shows that the scientific community in general has shown interest in this field of study.
Country analysis
Figure 2B shows the scientific production according to countries and distinguishes the 10 most important at the international level. India is the country that publishes the most studies, with 137 studies (26%), followed by China, Korea and Pakistan with 70 (13%), 27 (5%) and 25 publications (5%), respectively. In fifth place is Brazil as the representative of Latin America with a total of 18 publications, equivalent to 3%. As for the top 10 countries, seven are from Asia with a total of 295 publications corresponding to 53% of the total scientific production worldwide, one country from the European continent with 11 publications (2%), one from South America with 18 publications (3%) and finally one country from North America with a total of 11 publications (2%). These 10 leading countries in the scientific production of the subject under study are of great relevance since they produce 62% of the publications generated worldwide, a situation in which the countries belonging to the Asian continent stand out as important pillars for research in the area of MTIBAP.
Author analysis
Table 2 lists the 10 main authors who publish the largest number of articles related to the subject of this study; the classification is performed according to the total sum of publications in the WoS and Scopus databases, on the other hand, the number of citations and the H-index of the au thors are useful to determine the impact of productivity and scientific quality; the H index is the measurement of the scientific production of a researcher, determined by the number of articles and the number of citations from them (Alonso et al., 2009; Cascón-Katchadourian et al., 2020). The authors with the highest publication (six published articles) are Kim Sang-dal linked to Ye ungnam University - South Korea (903 citations and an H index of 15), Passaglia Luciane Maria Pereira from the Federal University of Rio Grande do Sul - Brazil (3475 citations and an H index of 25) and Yao Tuo from the Agricultural University of Gansu, China (379 citations and an H in dex of 9). It is relevant to mention that Pandey, Ashok linked to the Indian Institute of Toxicology Research - India, is placed at number nine in the list, according to Scopus with a number of citations of 46614 and with an H index of 99, warning that he has only four publications. It is also highlighted Maheshwari, Dinesh Kumar linked to Gurukul Kangri University - India, who is ranked number eight in the list and according to Scopus with a number of citations of 3469 and an H index of 34 with a total of five publications. It is highlighted that some authors have fewer publications than others, but their H index is higher, indicating that they are researchers with greater impact within the scientific community.
Table 2. Most relevant authors.
| Authors | Total publications | Country | WoS | Scopus | ||||
|---|---|---|---|---|---|---|---|---|
| Publications | Citations | H-Index | Publications | Citations | H-Index | |||
| Kim, Sang-dal | 6 | South Korea | - | - | - | 6 | 903 | 15 |
| Passaglia, Luciane María Pereira | 6 | Brazil | 3 | 2551 | 21 | 6 | 3475 | 25 |
| Yao, Tuo | 6 | China | - | - | - | 6 | 379 | 9 |
| Amaresan, Natarajan | 5 | India | 4 | 487 | 15 | 5 | 592 | 15 |
| Dastager, Syed Gulam | 5 | India | 5 | 1764 | 22 | 5 | 2101 | 24 |
| Hameed, Sohail | 5 | Pakistan | 5 | 1495 | 21 | 5 | 1773 | 23 |
| Khan, Mohammad Sayyar | 5 | China | 5 | 347 | 11 | 5 | 662 | 14 |
| Maheshwari, Dinesh Kumar | 5 | India | 5 | 2476 | 29 | 5 | 3469 | 34 |
| Pandey, Ashok | 4 | India | 4 | 38327 | 90 | 4 | 46616 | 99 |
| Castro, Paula M.L | 4 | Portugal | 4 | 2624 | 30 | 4 | 8648 | 52 |
Scientific journal analysis
Table 3 shows the 10 scientific journals with the highest number of publications on the subject of MTIBAP. The records are shown in relation to the Web of Science and Scopus databases, which have been used for this review, as well as the quartile, H-Index, impact factor, country and percentage of publications of the journals. The data show that of the ten journals with the highest impact, 40% are located in the Q2 category, the remaining 60% are distributed equally in 20% in each category Q1, Q3 and Q4 respectively; thus, the quartiles indicate the position of a journal in relation to all those in its area, ordered from highest to lowest impact using four quartiles. The journals with the highest impact will be the first quartile (Q1), the middle quartiles will be the second (Q2) and the third (Q3) and the lowest quartile will be the fourth (Q4). As for the journal with the most publications on the topic of study is Current microbiology from the United States, with a total of 15 publications, in second place is the journal microbiological research from Germany with 11 publications in total and in third position is the journal Frontiers in microbiology from Switzerland with a total of 10 publications; according to these data, the first three journals contribute 7% of the world's scientific production on MTIBAP. On the other hand, 50% corre sponds to European countries (Germany, the Netherlands and Switzerland), 30% to North Ameri can countries (United States and Canada) and the remaining 20% to India. According to Scimago Journal and Country Rank (SJR), the journal with the highest H-index is the journal Frontiers in microbiology with an H-index of 166, the journal with the best SJR index is also Frontiers in mi crobiology with 1.31; in addition, it is observed that some journals have fewer publications than others, but their H-index and SJR index is higher, indicating that they are journals with a high im pact on the scientific community.
Table 3. Scientific journal analysis.
| Journal | WoS | Scopus | Total | Percentage | Quartile | SJR (2021) | H- Index (SJR) | Country |
|---|---|---|---|---|---|---|---|---|
| Current microbiology | 15 | 15 | 15 | 3 % | Q3 | 0.48 | 97 | United States |
| Microbiological research | 11 | 11 | 11 | 2 % | Q2 | 1.11 | 94 | Germany |
| Frontiers in microbiology | 11 | 10 | 10 | 2 % | Q1 | 1.31 | 166 | Swiss |
| Journal of pure and applied microbiology | 6 | 10 | 10 | 2 % | Q4 | 0.21 | 20 | India |
| Biocatalysis and agricultural biotechnology | 4 | 9 | 9 | 2 % | Q1 | 0.64 | 46 | Netherlands |
| World journal of microbiology and biotechnology | NA | 9 | 9 | 2 % | Q2 | 0.67 | 98 | Netherlands |
| Canadian journal of microbiology | 7 | 7 | 7 | 1 % | Q2 | 0.55 | 100 | Canada |
| Journal of basic microbiology | 7 | 7 | 7 | 1 % | Q3 | 0.46 | 60 | Germany |
| Research journal of biotechnology | 7 | 7 | 7 | 1 % | Q4 | 0.13 | 18 | India |
| Biomed research international | 6 | 6 | 6 | 1 % | Q2 | 0.65 | 147 | United States |
Figure 3 shows the four relevant groups that make up the bibliographic study. In the first grouping, the network of co-citation of authors (A), this allows highlighting the most cited writers, in this case Glick Bernard, Schwyn Bernhard and Zhang Xiuhai are the most referenced. The second grouping, the network of collaboration of authors (B), highlights the work in 10 groups, the first group is the most significant composed of six researchers, which stand out Zhang Xiuhai, Khan Mohammad Sayyar and Xue Jing, since they have better literary relationship; the second group is constituted by three researchers Pandey Ashok, Dastager Syed and Deepa C. K, the most collaboration is between Pandey, Dastager; the third group consists of three authors, Liu Ke, Kloepper Joseph and Hu C.H, which have equal literary relationship. The rest of the groups con sist of two scientists each.
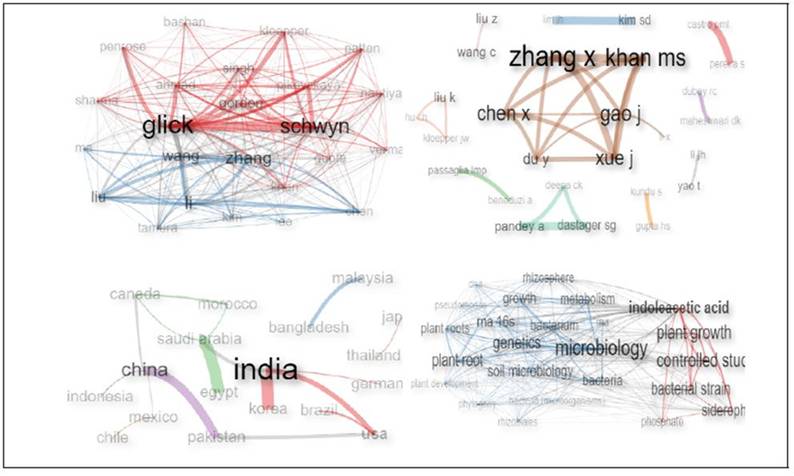
Figure 3. Elements of bibliometric analysis. (A) Author co-citation network, (B) author collaboration network, (C) cross-country collaboration network and (D) keyword co-occurrence network.
Now, the network of collaboration between countries (C) is the third constituent of the bibliometric analysis, showing that the country with the highest production of scientific articles about MTIBAP is India, with a marked collaboration with Korea, followed by China cooperating with Pakistan. Finally, the fourth group is shown, which consists of the network of cooccurrence of words (D), finding two broad groupings: the first in blue, the words microbiology, genetics, soil microbiology stand out; the second group in red, the words indoleacetic acid, plant growth and controlled study.
Network analysis
In this item, the most relevant studies on the subject were recognized. The documents with the most outstanding indicators were selected for analysis and organized using the metaphor of the tree of science. The 10 most significant (root), 10 pillars (trunk) and 40 recent documents (leaves). To establish the related research documents, the clustering algorithm was implemented according to Blondel et al. (2008), a simple method designed to extract community structures from large networks, allowing the visualization of distinct communities (clusters) in a network consisting of closely related nodes (Fig. 4).
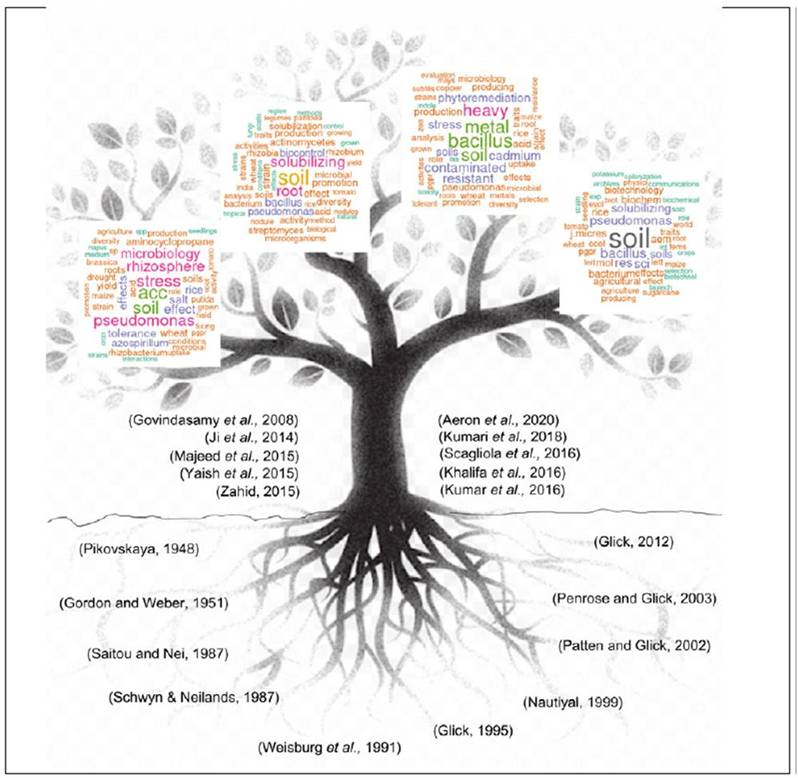
Figure 4 Diagram of the metaphor of the tree of science. The graphic description of the hegemonic authors (root), those who have consolidated the predominant research (trunk) and the trends with respect to the subject matter (leaves) is presented.
Root
The researches that make up the root of this bibliographic search are categorized as the studies with the highest indegree (group of most cited documents), establishing their superior and predominant condition. Thus, 10 publications were studied, cited as predominant according to the aforementioned. In this section, reference is made to methods for detecting certain compounds such as siderophores and indoleacetic acid (IAA); also procedures for isolating PGRP, accompanied by two literature reviews on microorganisms that facilitate plant growth. In addition, they re fer to techniques for the identification of phosphate solubilizing microorganisms. They include phylogenetic studies from the taxonomic point of view applied to these microorganisms and stud ies related to the improvement of root development in plants. These are specified below:
Before the implementation of molecular techniques such as polymerase chain reaction (PCR), the study of plant growth-promoting microorganisms was initiated using conventional methods as described below. In the middle of the 20th century, Pikovskaya (1948) was the first to develop a reliable and useful approach for the detection of phosphate solubilizing bacteria (PSB), which is currently applied and is based on visual detection of clarified zones in the culture medium, containing insoluble mineral phosphates. The performed approach is used by having 0.1 mL or 1 mL of suspension of a rhizosphere-associated soil sample serially diluted in sterile Pikovskaya (PVK) medium, supplemented with insoluble tricalcium phosphate as the sole source of phosphorus, dextrose and numerous inorganic components, for optical detection of clear zones in the medium. Also, the solubilization index (SI) was described by Pikovskaya (1948) as: SI=colony diameter (cm)+halozone diameter (cm)/colony diameter (cm). Years later, Gordon and Weber (1951) modi fied the ferric chloride-sulphuric acid method proposed by Tang and Bonner (1948) or the colorimetric estimation of indoleacetic acid (IAA), these changes produced greater stability, color, specificity and modification of the density more rapidly with respect to the variation in the concentration of IAA; they were able to conclude that by substituting sulfuric acid for perchloric acid in the Tang and Bonner reagent, the color intensity was improved and therefore it had the advantage of being a relatively sensitive method with greater specificity than the methods that had been used up to that time. On the other hand, at the end of the 80's Schwyn and Neilands (1987) developed as a research objective to describe a highly sensitive chemical method for the detection of siderophores based on their affinity for iron(III). Thus, they developed a global assay using Chromium Azurol S (CAS) and hexadecyltrimethylammonium bromide (HDTMA) as indi cators, based on the fact that CAS/HTDMA forms complexes with iron (III) to produce a blue color and when a strong chelator, such as a siderophore, suppresses the iron in the coloring com plex, its coloration varies from blue to orange.
According to the study of Saitou and Nei (1987) standardized a new method known as Neighbor-joining (NJ), which aimed to find pairs of operational taxonomic units (OTU) that decreased the total branch length at different stages of OTU clustering, from a star-shaped phylogenetic tree of any organism, which showed that the NJ method provided reliability and efficiency for the correct production of the tree topology. For their part, Weisburg et al. (1991) were among the pio neers in the use of molecular techniques, since they described a set of primers capable of initi ating enzymatic amplification, that is to say PCR, in a wide phylogenetic and taxonomic variety of bacteria, demonstrating the method for its use and a respective example; arguing that by means of oligonucleotides designed for genetic fractions, bacterial species considered difficult to culti vate or with the potential to be pathogenic can be studied.
In addition, Glick (1995) in his literature review noted the importance of considering and discussing the ways in which PGPR facilitate plant growth; the author described in his study the di rect mechanisms of plant growth promotion of PGPR and highlighted the relevance of the extensive biochemical and molecular studies carried out in symbiotic diazotrophs such as rhizobia, which have served as a starting point for understanding these mechanisms, due to the fact that rhizobia offer plants fixed nitrogen in exchange for carbon in photosynthesis; he also emphasized the processes of iron-siderophore bacterial complexes as a mechanism for obtaining iron from the soil, highlighting that without the presence of this process, plant growth would be severely lim ited; he also mentioned the effects of the synthesis of phytohormones, such as indoleacetic acid (IAA), an auxin phytohormone well known for stimulating rapid responses such as cell elonga tion, and participating in the processes of cell division and differentiation. On the other hand, Nautiyal (1999) formulated a defined medium for the detection of phosphate solubilizing micro organisms; these authors used the Pikovskaya medium (PVK) mentioned above, to perform the comparison, demonstrating that phosphate solubilization was three times more effective in the growth broth of the National Botanical Research Institute (NBRIP) compared to the PVK medium. For their part Patten and Glick (2002), demonstrated that the ipdc gene is essential to in crease the phytohormone indole acetic acid (IAA) of Pseudomonas putida from tryptophan, which has a significant impact on stimulating the growth of primary roots of seedlings, taking into account that they used PCR to isolate the gene of interest and carry out the research.
In the study of Penrose and Glick (2003), described a method to isolate and characterize plant growth-promoting rhizobacteria containing the enzyme 1-Aminocyclopropane-1-carboxylic acid (ACC deaminase) as they reduce ethylene levels, based on research by Glick (1995); this type of procedure allows other researchers to easily isolate new strains of PGPR containing ACC deaminase as an agricultural potential. According to the study of Glick (2012) continued his own re search cited above; he mentioned that soil bacteria in general and plant growth promoting bacteria (PGPB), produce cytokinins and gibberellins or both; he described that cytokinins (molecules that have the ability to promote cytokinesis or cell division in plants), have been detected in cells of Azotobacter spp., Bacillus subtilis, Pseudomonas fluorescens, Rhizobium spp., Pantoea ag glomerans, among others; he also concluded that the use of PGPB is recognized as an integral component of agricultural practices whose time has come and that the usefulness of these bacteria has been successfully demonstrated in growing countries around the world.
Trunk
In this section, the R studio program yielded 10 researches classified as the most representative in relation to the research topic. Thus, the concomitant documents in this item are classified as those whose betweenness or intermediation are the highest (authors who cite those of the root section and are cited in turn by the most recent scientific production, that is to say the clusters); in this way, research related to isolation and molecular identification techniques that allow charac terizing plant growth promoting microorganisms in cereals, vegetables, fruits, legumes and other crops such as palm are included.
In the study of Govindasamy et al. (2008), by PCR, cloned and sequenced the ACC deaminase (acdS) gene, managing to identify two bacterial isolates from wheat rhizosphere; thus, by phylogenetic analysis, the presence of Achromobacter sp. and Pseudomonas stutzeri was observed. Now, Ji et al.(2014) identified through specific primers of the nifH gene, two species of Paenibacillus, three species of Microbacterium, three species of Bacillus and four species of Klebsiella, which were isolated from 10 rice fields, in addition, they demonstrated that the characterized diazotrophic endophytic bacteria could be used as plant growth promoters and induce fungal resistance in plants. Subsequently, Majeed et al.(2015) managed to obtain nine bacterial isolates, of which seven produced indole-3-acetic acid; seven were nitrogen fixers and four were able to solubilize inorganic phosphate in vitro; they also evidenced genotypically four different morphotypes, based on IGS-RFLP (Intergenic Spacer - Restriction Fragment Length Polymor phism) fingerprinting and by 16S rRNA sequencing, identified Stenotrophomonas spp. and an other bacterial isolate with genetic traits equal to Acetobacter pasteurianus and Stenotrophomonas; affirming the potential of PGPR in the early growth of wheat (Triticum aestivum L.). In addition, Scagliola et al.(2016) isolated, characterized and molecularly identified bacterial strains from the rhizosphere of barley (Hordeum vulgare L.) and tomato (Solanum ly copersicum L.), previously cultivated under hydroponic conditions; isolation was performed us ing the standard plate dilution technique with nutrient agar supplemented with 0.1% (w/v) cyclo heximide as fungal inhibitor; for PCR amplification of the 16s rDNA gene, the template of the genetic material was extracted by heat shock and using the universal primers 357f and R1401, amplicons of 1,060 bp were evidenced; the results showed that of the 800 isolated strains only 80 excreted siderophores, of which belonged to 12 different genera, the most representative being Pseudomonas (approximately 80%), but also genera such as: Enterobacter, Azotobacter, Stenotrophomonas, Chryseobacterium and Rhizobium.
In another study, Yaish et al. (2015) isolated and characterized endemic endophytic bacteria from date palm (Phoenix dactylifera L.) and identified some of the mechanisms that these bacteria can use, to facilitate the growth of this plant in saline environments; for the isolation of bacte rial endophytes they used two techniques; in the first, root tissues were extracted using Ringer's solution and then seeded directly on agar media rich in different components and in the second, aliquots of Ringer's solution were taken, extracted as inocula from the selective media in sequen tial procedures, for the isolation of ACC deaminase-producing strains; as for molecular identifi cation, they amplified the 16S rRNA gene using primers 27F and 1492R for bacteria; 16S rRNA gene sequence analysis led to the isolation of bacterial strains using the first method related to the genera Bacillus, Chryseobacterium, Paenibacillus, Rhodococcus and Staphylococcus, where Bacillus was dominant, and the second method led to the isolation of endophytes related to the genera Achromobacter, Acinetobacter, Escherichia, Enterobacter and Klebsiella, where Enter obacter was dominant. On the other hand, Zahid et al. (2015) isolated and identified PGPR asso ciated with maize (Zea mays L.) crops in Pakistan; for molecular identification and authentica tion, eubacterial primers fD1 and rD1 corresponding to the 16S rRNA gene of Escherichia coli were applied; the results of the application of 16s rRNA gene sequencing and phylogenetic analy sis evidenced that the isolates belonged to bacteria of the genus Pseudomonas and Bacillus.
So, the study of Kumar et al. (2016) designed an investigation to isolate and characterize PGPB (Pseudomonas spp.) from the rhizosphere of common bean (Phaseolus vulgaris L.), from which they selected 8 isolates of Pseudomonas (PPR1-PPR8) and carried out the amplification of the PPR8 16s rRNA gene by PCR, using the universal eubacterial primers fD1 and rD1; the sequence of the gene used was aligned for similarity using the BLAST program of the GenBank database (NCBI) and thus, the authors obtained as a result of the sequencing of the 16s rRNA gene of Pseudomonas sp. PPR8, a sequence of 904 bp showing 100% similarity with Pseudomonas sp. BP-1 and 99% similarity with Pseudomonas sp. MBEA06 and Pseudomonas sp. HNS021. For their part, Khalifa et al. (2016) also performed 16s rRNA gene amplification using the universal primers 27F and 1492R, from a DNA template of an endophytic Enterobacter cloa cae (MSR1) strain, isolated from inside roots of alfalfa (Medicago sativa) plants; the 16s rRNA gene sequencing results exhibited 99% homology with E. cloacae subsp. cloacae strain DSM 30054T, E. cloacae strain 5621A, E. cloacae strain AB6, and E. cloacae strain E717. In the study of Kumari et al. (2018) isolated and identified Pseudomonas spp., Bacillus spp., and Acinetobacter spp., by biochemical analysis and 16S rDNA gene sequencing, which were isolated from the rhizosphere of mung bean plants; they even demonstrated that all bacterial strains turned out to be PGPR, capable of producing indole-3-acetic acid and solubilizing phosphate. On the other hand, Aeron et al. (2020) identified Stenotrophomonas maltophilia by 16S rRNA sequence, which was recognized as a non-rhizobial endophytic bacterial strain, isolated from the root nodules of the medicinal legume Mucuna utilis var. capitata L.; demonstrating that it promotes plant growth di rectly and indirectly.
Leaves
From the bibliographic review, four perspectives or clusters were characterized, in this thematic under study, the most relevant lines of research in the area addressed are manifested. These are specified below:
In this first cluster(Fig. 5), there are the investigations that mention PGP bacterial genera such as Bacillus, Pseudomonas, and also the importance of the identification of microorganisms with mineral solubilizing potential, because often the availability of these nutrients in the soil is limited; based on this Chakdar et al. (2018) carried out the isolation, identification and molecular characterization of mineral phosphate solubilizing bacteria from thermitorial soils of Sanjivani Island, India; the results of molecular amplification of 16s rRNA gene with primers (16S F27 and 16S R1525) demonstrated highly efficient isolates, such Pantoea sp. A3, Kosakonia sp. A37, who produced and increase of 37 and 53% of tomato root length; In addition, Bacillus sp. AH9 was identified, this isolate showed the highest phosphate index and the highest solubilization efficiency in Pikovskaya Agar. In the study of Mushtaq et al. (2019) developed the screening of bac terial endophytic isolates through citrus leaves for plant growth promoting activities using molecular characterization by using bacterial primers (27F and 1492R); the results evidenced, that out of 16 isolates characterized, Pseudomonas aeruginosa, Pseudomonas sp., Brevibacterium borstelensis, Enterobacter hermachei, Klebsiella pneumoniae, Staphylococcus haemolyticus, Bacillus subtilis, Psychrobacterium pulmonis, Bacillus megaterium, Proteus mirabilis and Bacillus cereus, showed phosphate solubilization activity. For their part, Jana and Yaish (2020), iso lated a rhizobacterium from date palm grown in salinity and by 16s rRNA gene amplification, us ing genomic DNA and universal primers 27F and 1492R, identified Enterobacter cloacae SQU-2, which in addition to the production of siderophores and indoleacetic acid (IAA), showed zinc, phosphorus and potassium solubilization characteristics. For their part, Lelapalli et al. (2021) iso lated and performed molecular characterization using the universal primers described by Weis burg et al. (1991), finding that Lysinibacillus pakistanensis PCPSMR15, is a potential bacterium of the rice rhizosphere, capable of solubilizing inorganic phosphorus from insoluble compounds and promoting plant growth in terms of root, shoot and leaf length.
Liu et al.(2020) isolated and identified a plant growth-promoting rhizobacterium from root soil samples of rice plants; the researchers used 16s rRNA gene sequencing and confirmed the close relationship with 99.93% similarity to Bacillus methylotrophicus (DD-1), an isolate that exhibited potassium solubilizing activity, production of indoleacetic acid, gibberellic acid, siderophores and increased growth parameters in dry weight, number of tillers, root and shoot length compared to plants not treated with the DD-1 isolate. In turn Zhu et al. (2020) also isolated and identified PGPR in rhizosphere soil samples from rice plants; they performed molecular identification for amplification and sequencing using the primers 27F and 1492R, but also primers for gyrA and gyrB, specific genes of the auxin-producing rhizobacterium Bacillus mega terium; the authors concluded that, of the thirteen isolates from the rice rhizosphere, three strains, B. megaterium, B. subtilis subsp. subtilis and B. subtilis were auxin-producing and salt-tolerant strains. In addition, Fasusi et al. (2021) performed genomic sequencing and PCR amplification with the universal primer for bacteria 341F and 907R of the 16s rRNA gene; finding isolates with potential plant growth-promoting activities in rhizospheric soil samples of maize (Zea mays L.) and soybean (Glycine max), identifying Bacillus spp. (80.77%), Rhodocyclaceae (3.85%), Ente rococcus spp. (3.85%), Massilia spp. (3.85%) and Pseudomonas (7.69%); the bacterial isolates possessed antifungal activity against Fusarium graminearum and promoted the development of corn and soybean seeds under laboratory conditions.
In another study, Gu et al. (2020) isolated and characterized at the molecular level, employing sequencing and amplification of 16s rDNA using 27F and 1494R oligonucleotides in rhizospheric soil samples of tobacco plants; strain S58 identified as Pseudomonas mediterranea, showed significant antagonism against plant pathogenic fungi and bacteria and efficiently controlled acute eyespot of wheat. For their part, Aldayel and Khalifa (2021) identified Pseudomonas monteilii in rhizospheric soil samples of tomato (Solanum lycopersicum L.) with 99.86% homologous regions by sequencing the 16s rRNA gene and applying the universal primers 27F and 1494R; the results evidenced that this strain significantly promoted primary root length, stem and leaf number in Eruca sativa (arugula) and Raphanus sativus (radish), growing under 1,000 mg kg-1 salt stress conditions. Finally, Aloo et al. (2020), investigated the diversity of cultivable rhizobacteria asso ciated with potato (Solanum tuberosum L.) in Tanzania; of the 152 isolates obtained, 52 of them were selected for molecular identification based on preliminary screening for PGP activities; thus for PCR amplification, primers 27F and 1492R were used; The results showed that the isolates were all Gammaproteobacteria, belonging to four families (Enterobacteriaceae, Yersiniaceae, Pseudomonadaceae and Morganellaceae) and nine genera (Enterobacter, Klebsiella, Serratia, Pseudomonas, Morganella, Buttiauxella, Pantoea and Cedecea).
In this second cluster (Fig. 6), it refers that currently agricultural practices have been revolutionized by microbiology, in search of sustainable agriculture by means of microorganisms with the potential to favor the factors or conditions that plants require to grow in a better way. In the study of Saengsanga (2018) molecularly identified Enterobacter sp. and Bacillus sp. strains isolated from the rhizosphere of early Thai jasmine rice plants with the ability to produce high concentrations of indoleacetic acid (IAA). On the other hand, Khezrinejad et al. (2019) characterized plant growth promoting rhizobacteria (PGPR) in sunflowers by phenotypic analysis and rep-PCR method; obtaining strains of Pseudomonas fluorescens, P. aeruginosa, P. geniculata, Bacillus subtilis, B. pumilus, Stenotrophomonas maltophilia and Brevibacterium frigoritolerans; in addition, by means of experimentation, they showed that the variability of microbial species obtained as PGPR improved the morphophysiological characteristics of sunflower shoots and roots. According to the study of Helal et al. (2022) the authors proposed a hypothesis on the effect of rhizocompetence in the rhizosphere of tomato, based on bacterial isolates from different microhabitats of this plant in different soils, identifying the genera Pseudomonas, Bacillus and six other bacteria by BOX-PCR; concluding that soil type and plant sphere can influence both genotypic diversity and rhizocompetence capacity of the same bacterial species.
In another research, Hyder et al. (2020) isolated and identified Pseudomonas putida, P. libanensis, P. aeruginosa, Bacillus subtilis, B. megaterium and B. cereus from chili rhizosphere by 16S rRNA sequence analysis, where it demonstrated the fundamental function of these microorganisms obtained for the antagonism of a fungus, in addition, their ability to improve the growth characters of the chili plant. For their part, Alotaibi et al. (2022) demonstrated that the combination of PGPR variety and petroleum hydrocarbon degradation (PHC) potential of bacteria contribute in the positive effect on canola shoot length growth and phytoremediation capacity to PHCs, where they were able to prove the mechanisms of Pseudomonas plecoglossicida, Bacil lus and other microbial species to perform this type of beneficial processes, which could lead to the development of innovative bacterial inoculants for plants to remediate contaminated soils. Taking into account Andleeb et al. (2022), they conducted a research based on the evaluation and identification of plant growth promoting bacteria from the digestive tract of Eisenia fetida (Striped Red Worm) achieving the characterization of Bacillus mycoides, B. aryabhattai, B. megaterium, Staphylococcus hominis, B. subtilis, B. spizizenii, B. licheniformis, B. mojavensis, B. toyonensis, B. anthracis, B. cereus, B. thuringiensis and B. paranthracis as producers of siderophores, indoleacetic acid, and other compounds, in addition to the ability to solubilize phosphate, demonstrating the ability of most strains of the genus Bacillus isolated to be used as potential microbial biofertilizers to increase crop production.
On the other hand, in agriculture, efforts have been made to select plant growth-promoting bacteria resistant to heavy metals to improve the efficiency of phytoremediation and in turn increase crop production. Accordingly, Fan et al. (2018) isolated endophytic bacterial strains from root nodules of Robinia pseudoacacia, grown in a Pb-Zn mining area; demonstrating that some of these microorganisms had the ability to resist Cd, Zn, Pb and Cu, also the biological activity to produce indole-3-acetic acid, siderophores and carry out 1-aminocyclopropane-1-carboxylate deaminase activity, where Mesorhizobium loti HZ76 was the best strain to promote plant growth and perform phytoremediation processes. Likewise, Wu et al. (2019) also characterized heavy metal resistant plant growth promoting bacteria from acid mine drainage contaminated crop soil, which evaluated plant growth, nutrient uptake, antioxidant enzyme activities, and plant growth promoting characteristics were determined; thus, they identified strains S6-1 of Burkholderia sp., and S2-3 of Pseudomonas sp. demonstrating higher resistance and activity as PGPs. On the other hand, Idaszkin et al. (2021) isolated bacteria from the rhizosphere of the halophyte plants Spartina densiflora and Sarcocornia perennis in a polluted marsh, obtaining 60 strains, of which 25% of them produced siderophores, 16% were able to solubilize phosphate, 11% were able to produce auxins and 7% chitinase; evidencing that bacteria isolated from the rhizosphere of S. densiflora demonstrated higher PGP properties related to heavy metal phytostabilization. Finally, Bennis et al. (2022), aimed to characterize plant growth-promoting rhizobacteria from the rhizo sphere of naturally grown R. pseudoacacia plants grown naturally in mining tailings contami nated with Pb and Zn, obtaining as a result the alteration of the properties of the three isolated PGPs when different concentrations of Pb or Zn were added to the culture medium; as for the analysis of the 16S rRNA gene of each strain, it was evidenced that strains 7MBT, 17MBT and 84MBT had 99.34, 100 and 99.72% of similarity with con Priestia endophytica, B. pumilus NBRC 12092 T and B. halotolerans NBRC 15718 T, respectively.
In the third cluster (Fig. 7), according to its great relevance and repeated application, additional studies are mentioned, related to the isolation and molecular identification of the bacterial genera Pseudomonas and Bacillus that present plant growth promoting mechanisms; also, the capacity of PGPB to solubilize insoluble minerals such as phosphates and potassium, attributed as vital microbial functions for plant development, is mentioned. Accordingly, Ong et al. (2018) characterized diazotrophic bacteria present in saline soils, plant rhizosphere and plant roots, amplifying by PCR employing primers (polF, polR) and (27F, 1492R) targeting nifH genes and 16S rRNA gene respectively; the results indicated that, out of 147 isolates, 10 demonstrated the best characteris tics related to plant growth mechanisms, identified as strains AS6. 3ES, SDSO11.2 and AS6.3ES, with 99, 100 and 99% sequence similarity to Pseudomonas sp., Pseudomonas plecoglossicida strain LJ3 and several strains of Pseudomonas putida, respectively, which were positive for ACC deaminase activity, IAA production, siderophore synthesis and phosphate solubilization. Years later, Singh et al. (2020) sequenced and performed molecular identification of three PGPRs based on 16s rDNA gene sequence and PCR amplification by primers 27F and 1492R; the three isolates were identified as Burkholderia arboris, Pseudomonas aeruginosa and Acinetobacter baumannii and showed multiple PGP activities in vitro, for example, P. aeruginosa showed maximum IAA production and B. arboris showed maximum mineral solubilization and production of siderophores and ammonia. According to the study of Leontidou et al. (2020), after molecular identification through sanger sequencing and amplification of the 16s rRNA gene with Bakt_341F and Bakt_805R primers in samples of wild plants and a tomato cultivar, the presence of Pseudomonas (48%) and Bacillus (20%), as well as Acinetobacter, Pedobacter, Pantoea, En terobacter, Luteibacter, Lysobacter and Chryseobacterium was determined; In addition, the iso lates of Pseudomonas sp. showed genes related to possible PGP and included the gdh and ppq genes encoding glucose dehydrogenase and pyrroquinoline quinone synthase respectively, se quences that are associated with the production of gluconic acid, a substance involved in the solu bilization of inorganic phosphate.
On the other hand, Singha et al. (2017) revealed the presence of nitrogen fixation genes (nifH) and nodulation genes (nodC) in isolates selected using fresh root nodules from pigeon pea (Cajanus cajan) and Lablab purpureus plants; for the molecular identification of the nifH genes they used the primers zehrf and zehr, while for the identification of the nodC genes they used the primers nodC1 and nodC2, likewise, they performed PCR-RFLP of the 16s rRNA gene applying the primers 27F and 1492R, thus obtaining that all isolates belonging to the genera Rhizobium, Mesorhizobium and Burkholderia were able to solubilize phosphates confirmed by a zone of clarification in the Pikovskaya agar medium. For their part, Malisorn et al. (2020), also amplified a DNA fragment by PCR, using the 16s rRNA gene using primers 27F and 1492R, a technique from which they identified 20 isolates obtained from legume nodules and rhizospheric soil samples, corresponding to the genera Rhizobium, Ochrobactrum, Pseudomonas and Acinetobacter. The results of the plant promotion mechanisms showed that the isolate SN-5 Pseudomonas geniculata showed the highest production of IAA and the highest rate of phosphate and zinc solu bilization. In the study of Mghazli et al. (2022), conducted an investigation with the objective of isolating, characterizing and identifying autochthonous bacterial strains in vitro, for the best per forming strains in plants, they performed sequencing and PCR amplification of the 16S rRNA gene using the primers 27f/1492r and fD1/rD1; the results of molecular identification reflected that the 41 isolates, corresponded to the genera Microbacterium, Bacillus, Acinetobacter, Agro coccus, Brevibacterium, Neobacillus, Paenibacillus, Peribacillus, Pseudarthrobacter, Stenotrophomonas and Raoultella; all isolates were able to tolerate up to 5% NaCl; Bacillus paramycoides showed the best catalase activity, positive reaction to cellulase synthesis and the highest hydrogen cyanide (HCN) production activity, a compound known for its ability to control and counteract plant pathogens.
Javoreková et al. (2021) identified PGPRs of the genera Azospirillum, Azotobacter, Arthrobacter, Bacillus, Clostridium, Enterobacter, Pseudomonas and Serratia by molecular analysis of the 16S rRNA gene using primers 27F and 1492R; isolates of which, the bacterial strains B. altitudinis, B. aryabhattai and B. megaterium, showed the best PGPR properties, thus representing an alternative for plant growth promotion in maize (Zea mays L.). Similarly Umapathi et al. (2022), conducted an investigation to identify drought-tolerant endophytic bacteria and to know their PGP effect on sorghum plants (Sorghum bicolor L.); to meet this objective, they isolated and identified bacteria of the genus Acinetobacter pittii, Bacillus licheniformis, Bacillus sp., Pseudacidovorax intermedius and Acinetobacter baumannii by 16S rRNA sequencing; they selected the strains A. pittii, Bacillus sp. and P. intermedius, to evaluate growth on sorghum seedlings, the results concluded that Bacillus sp. and P. intermedius improved growth character istics: root length, shoot length, plant vigor index and total dry matter production, and protected the seedling against water stress.
For their part, Bhutani et al. (2022) recognized that the bacterial endophyte Bacillus licheniformis isolated from nodules and roots of Vigna radiata (mung bean), this strain was the only one of the 46 isolates that managed to grow at a salt concentration of 15%, so that after determining its molecular identity mediated by 16S rDNA sequencing, it was declared positive for ACC deaminase activity, IAA production, siderophores, ammonia, hydrogen cyanide (HCN), which means a potential isolate to promote plant growth in soils where there is a seasonal variation of salinity as a consequence of climate change. Also, Gohil et al. (2022) isolated and identified Bacillus sp. PG-8 from fermented Panchagavya, (mixture among five cow products including manure, urine, milk, rennet and butter), by applying 16s rRNA gene sequence using universal primers 27F and 1492R; due to its PGB characteristics such as IAA production, gibberellic acid, exopolysaccharide production, ammonia synthesis and phosphate solubilization, strain PG-8 was tested on Arachis hypogea (peanut), showing a germination rate of 70% in seeds treated with this inoculum, optimizing growth compared to the control and considerably improving root length, number of root hairs, number of leaves and leaf area.
In this last cluster (Fig. 8), the influence of biotic or abiotic factors is analyzed; however, some microorganisms have the capacity to diminish the negative effects of stress, increasing plant tolerance to these factors. South et al. (2021) conducted a study based on the search for PGPR, which could improve the performance of ornamental plants at greenhouse level, with a lower amount of fertil izer, considered as an abiotic stress factor; so they genetically identified Caballeronia zhejian gensis C7B12, considering it as a potential PGPR under the given conditions of stress. On the other hand, Panigrahi et al. (2020) performed morphological, biochemical, and 16S rRNA gene sequencing characterization of an endophytic bacterium, identified as Enterobacter cloacae, where they determined that this isolated strain, was able to tolerate 9% NaCl and grow at 54 °C under osmotic stress in the medium, in addition to significantly improve the growth of four crop plants. However, Saravanakumar (2019) states that there are other types of mechanisms to benefit plants in the face of a variety of stress factors, where microorganisms may contain genes that fa vor the production of enzymes as a defense against adverse compounds that harm plant health, in this case 1-aminocyclopropane-1-carboxylic acid deaminase (ACCD), produced by microorgan isms, to promote the growth of plants that are influenced by environmental stress due to high eth ylene accumulation.

Figure 8. Cluster 4, most mentioned words: Pseudomonas, Rhizosphere, ACC (1 - Aminocyclopropane - 1 - carboxylate (ACC) deaminase), Stress.
There are other abiotic stress factors such as temperature; thus, Vega-Celedón et al. (2021) affirm that cold is one of the factors that minimizes plant growth, therefore, these authors aimed to isolate and characterize PGPB psychrotolerant of wild flora, which have the ability to protect and promote plant growth at low temperatures; thus, they identified the 16 rRNA gene of the isolated strains, and formulated PGPB consortia; obtaining that the bacterial consortium composed of Pseudomonas sp. TmR5a and Curtobacterium sp. BmP22c were the best, since it showed the presence of ACC deaminase and ice recrystallization inhibition activities. According to Goyal et al. (2022), they argue that PGPBs must have dual functional capacity to be implemented in arid crops; firstly the expression of ACC deaminase and secondly, the ability to tolerate higher tem perature and salt concentration; so they isolated PGPB from the rhizosphere of Cyamopsis tetragonoloba, and sequenced the 16S rRNA gene of the microorganisms, identifying Pseu domonas, Enterobacter and Stenotrophomonas, they were ACC deaminase positive, thermohalotolerant and drought tolerant successfully. Govindasamy et al. (2022) presented the first report of isolation and characterization of cactus endophytic actinobacteria, where they identified strains of Streptomyces sp. by 16S rRNA gene sequencing and phylogenetic analysis, which were noted for their plant growth promotion such as ACC deaminase activity; even, the potential to improve crop plant growth under abiotic stress conditions such as drought. In the same year, Gao et al. (2022) aimed to isolate and identify endophytic antagonistic bacteria from lily roots, evaluating their antifungal activity and plant growth promotion characteristics; they recognized Bacillus halotolerans by 16S rRNA sequence, demonstrating ACC deaminase activity, salt and drought stress tolerance in an in vitro experiment, in addition to other mechanisms that contributed to plant growth promotion in this research.
On the other hand, Jhuma et al. (2021) confirm that the application of PGPR is a strategy of great viability to counteract biotic and abiotic stresses; these authors isolated 65 endophytic bacteria from the roots of healthy Oryza sativa, naturally grown in a saline environment; thus, by ana lyzing ribosomal DNA, they identified four genomically diverse groups, which were Enterobacter, Achromobacter, Bacillus and Stenotrophomonas, evidencing the tolerance of these microorganisms to NaCl ranging from 1.37 to 2.57 mol L-1 in nutrient agar medium; furthermore, under a salt stress of 200 mmol L-1 in vitro, the isolated strains of 108 CFU/mL demonstrated competitive production of exopolysaccharides (EPS). However Djebailiet al. (2021), analyzed fourteen isolates for phosphate solubilization, indoleacetic acid, hydrocyanic acid and ammonia production under different salt concentrations, showing that all isolates had halotolerant ability, improved morpho-biochemical parameters of durum wheat plants, and also 86% had 1-aminocyclopropane-1-carboxylate deaminase activity. According to Li et al. (2022), one of the main fac tors that threaten tall fescue (grasses) growth and turf quality is soil salinity; however, 10 genera were obtained from isolation, demonstrating the importance of Bacillus zanthoxyli and Bacillus altitudinis as promoters of tall fescue growth, based on salt tolerance.
CONCLUSION
Since 2012, there has been a significant increase in scientific production on the subject of MTIBAP, due to the need to identify and apply plant growth promoting bacteria on different agricultural crops, as an alternative to the use of agrochemicals. Regarding the molecular biology techniques used by different authors, sequencing and PCR amplification are the most frequent to identify and characterize the microorganisms that have shown a better response in in vitro and ex vitro research, using the universal eubacterial primers 27F and 1492R or 1494R.
Since the creation of PCR and sequencing techniques, greater speed, precision, sensitivity and specificity have been obtained for the identification of microbial genera isolated from agricultural soils, with potential for application as biofertilizers. The diversity of microorganisms isolated from the soil have been studied for their capacity to favor plant growth, mainly by solubilization mechanisms, production of siderophores and phytohormones and expression of ACC deaminase; the genera that have been most identified molecularly by using these mechanisms are Bacillus, Pseudomonas, Enterobacter and Acinetobacter.
In the purpose of continuing to address the subject and apply future research, it is suggested to continue developing the perspectives alluded to by the authors (Tab. 4); some of these are specified below:
Table 4. Perspectives.
| Perspective | Topic | Reference |
|---|---|---|
| Solubilization, Bacillus, Pseudomonas, root | Bacterial endophytes could be more useful in controlling diseases of field crops | Mushtaq et al. (2019) |
| B. subtilis subsp. subtilis and B. subtilis show potential for inducing salt tolerance in alfalfa plants | Zhu et al. (2020) | |
| Apply the use of L. pakistanensis as a biofertilizer and conduct further research due to its enzymatic characteristics and eventual commercial applications | Lelapalli et al. (2021) | |
| Bacillus, soil, metal, resistance, heavy | Heavy metal contaminated areas can be biologically remediated by the interaction between heavy metal resistant rhizobia and Robinia pseudoacacia. | Fan et al. (2018) |
| Importance of native rhizobacteria for the control of soil oomycetes and the potential use of Bacillus and Pseudomonas spp. in the development of biofertilizers and biofungicides | Hyder et al. (2020) | |
| Several strains obtained by these authors have the potential to be biofertilizers, demonstrated by the in vitro activities performed, as well as their high capacity of rhizocompetence, which is influenced by the type of soil and the plant | Helal et al. (2022) | |
| Soil, Bacillus, Pseudomonas, solubilization | Conduct future research from a more holistic approach of PGP characteristics and isolation of microbial consortia as an agricultural application in unfa vorable environments | Leontidou et al. (2020) |
| Continue to test in vivo bacterial strains of B. megaterium, B. altitudinis and B. aryabhattai to promote maize growth without or with minimized use of in dustrial fertilizers and pesticides | Javoreková et al. (2021) | |
| Molecular mechanisms for adaptation to salinity due to climate change can be understood using the halotolerant bacterium Bacillus licheniformis | Bhutani et al. (2022) | |
| Pseudomonas, rhizosphere, ACC (1-Aminocyclopropane-1-carboxylate (ACC) deaminase), stress | E. cloacae strain MG001451 could be an invaluable resource for applied agriculture, reducing the use of synthetic fertilizers and contributing to sustainable agriculture with further scientific research | Panigrahi et al. (2020) |
| Further studies should be conducted to evaluate the possibility of producing biofertilizers with strains S. iakyrus G10, S. ambofaciens J27. S. xantholyticus K12, making consortia to evaluate possible synergistic effects, and to investigate the effectiveness of the formulations in open field experiments and on differ ent crops for the development of biofertilizers useful to overcome high salinity in soils | Djebaili et al. (2021) | |
| Bacillus halotolerans LBG-1-13 can be considered a potential biocontroller and biofertilizer in lily, due to its antifungal capabilities, drought and salt tolerance, and production of ACC deaminase | Gao et al. (2022) |