Introduction
Panicum coloratum L. (subfamily Panicoideae, tribe Paniceae) is a warm season perennial grass originating in Africa and introduced to Argentina around 1990 to be used as forage (Tischler and Ocumpaugh 2004). It is referred to in the literature as an allogamous cross-pollinated species (Pritchard and De Lacy 1974), and chromosome numbers are variable, although the most frequently reported is the tetraploid 2n=4X=36. The species is represented in Argentina by 2 varieties: var. makarikariense Gooss. (Goossens 1934) and var. coloratum (Bogdan 1977). It has been introduced to the country in multiple events that have been partially documented; collections are scarcely preserved in active genebanks (Armando et al. 2013). There has been renewed interest in the species recently following the relocation of livestock production to less productive areas and the subsequent need to produce forage of reasonably good quality and quantity in these situations (Rearte 2007; Manuel-Navarrete et al. 2009). As a result breeding efforts have been initiated in Argentina, since P. coloratum is one possible option for overcoming forage shortage in areas with climatic and edaphic constraints.
Breeding of perennial grasses is a lengthy process that involves several phases, such as gathering basic information about aspects of reproductive biology and cytology. It is essential to acquire a range of genetic material by compiling germplasm sources from different collections and evaluating it prior to performing selection and breeding (Vogel and Burson 2004). Breeding efforts with P. coloratum var. coloratum have led to cultivars with increased seedling weight (Hussey and Holt 1986) and the development of several lines with improved agronomic characteristics such as good seed retention (Young 1986), seed dormancy (Tischler and Young 1983) and mesocotyle length (Tischler and Voigt 1995). Although the species shows promise for use in subtropical areas, several traits still need to be improved to increase its adoption by ranchers. In fact, only a single cultivar, cv. Klein, is currently available to farmers in Argentina, and seed quality is frequently diminished by shattering problems.
Prior to commencing breeding activities, this study aimed to: a) evaluate the genetic diversity in the germplasm collection at the Instituto Nacional de Tecnología Agropecuaria (INTA) in Argentina, using inter-simple sequence repeat markers (ISSRs) and agronomically relevant morphological characters; b) compare the variability within and between accessions in the collection and with the commercial cv. Verde Kleingrass; and c) harmonize configurations rendered by the 2 types of traits and use them for the simultaneous characterization of accessions to be further used in conservation and breeding.
Materials and Methods
Plant material
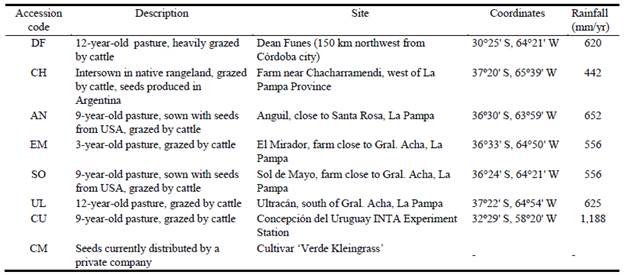
The germplasm of P. coloratum var. coloratum used for this research was collected in 2006 from 7 different locations in 3 provinces in Argentina: Córdoba (accession DF), La Pampa (accessions CH, AN, EM, SO, UL) and Entre Rios (accession CU). The collection is preserved at the INTA Rafaela Experimental Station (31°11'41" S, 61°29'55'' W; 957 mm mean annual precipitation). Collection sites differed in terms of soil characteristics, annual rainfall, original seed source, grazing management, etc. (Table 1). Individual accessions in the collection consisted of 32 plants in a common garden, separated at 0.60 m from one another. Different accessions were separated by a distance of 15 m to reduce the possibility of cross-pollination. The collection also included a plot of individuals from the commercial cv. Verde Kleingrass (CM; from here on referred to as 'cv. Klein'). Plant morphological characterization was performed between January and April 2011, during which 821 mm accumulated rainfall was recorded, a figure well in excess of the historical precipitation for that period (485 mm); mean temperature was 23.4 °C. The soil at this location is classified as Argiudoll (loamy, approximately 3% organic matter content, neutral pH, medium nutrient content and very low electrical conductivity).
Morphological and agronomic evaluation
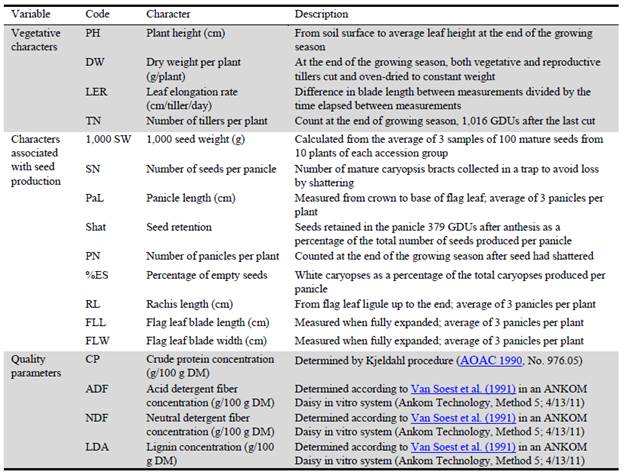
Measurements were performed on 10 individual plants for each accession, giving a total of 80 plants. A group of 17 morphological variables were recorded, related to forage production, forage quality and characters associated with seed production. Descriptions of variables and abbreviations used are displayed in Table 2. All observations were performed on adult plants. Three tillers per plant were tagged before starting measurements to calculate leaf elongation rate (LER). After an initial harvest at 15 cm height, a single growing leaf per tiller was measured every 5-7 days from 25 January to 17 February 2011. Daily growing degree units (GDUs) were calculated as
GDU = [(Daily maximum temperature - Daily minimum temperature)/2] - Tb
where Tb was set to 10 °C base temperature (Ferri et al. 2006). Total aerial biomass was obtained by weighing the total biomass produced after 1,016 GDUs from the initiation of the measurements. The dried (60 °C for 72 hours) material was further milled using a 2 mm mesh before determination of quality parameters as described in Table 2.
Table 2 Morphological traits used to characterize Panicum coloratum var. coloratum and sampling methods.
Molecular evaluation
Genomic DNA was determined on 3-5 young fresh leaves from each of the plants used for morphological evaluation following the protocol CTAB described by Sambrook et al. (1989). The genomic DNA quality was evaluated by electrophoresis in agarose 0.8% gels with ethidium bromide (0.5 ng/mL) and 0.5X TBE buffer (45 mM Tris base, 45 mM boric acid, 1 mM EDTA pH 8.0) in 1X TBE buffer at 70 V during 1 h, and visualized under UV light and photographed with the G-box (Syngene®). Specific primers from 5.8s ribosomal gene were designed and used to confirm that the extracted DNA actually belonged to P. coloratum.
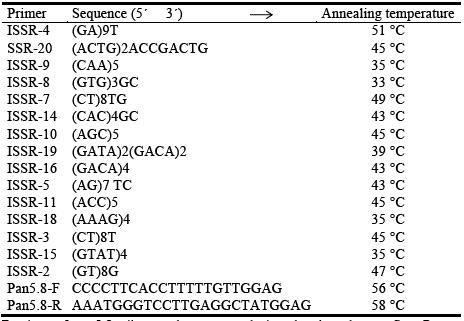
PCR reactions were performed in a Gene Amp PCR System 9700 thermocycler. Fifteen ISSR primers were tested for amplification (Garayalde et al. 2011). The reactions were performed in a final volume of 25 mL of a mixture containing buffer [75 mM Tris-HCl of pH 8.8, 20 mM (NH4)2SO4, 0.01% (v/v) Tween 20, 2 mM MgCh, 0.2 mM of each dNTP, 0.2 (jJVI of primer, 0.05 U/pL of Taq polymerase (Invitrogen) and ~15 ng/fiL of genomic DNA (dilution 1/100 of DNA stock genomic)]. Simultaneously with each PCR reaction, a mock sample was added plus 2 controls, consisting of product amplifications of 2 individuals (CH9 and CH10) for each primer (with the aim of revealing the presence of non-specific bands). The program used in the PCR reactions was the same for all 15 primers tested and consisted of an initial period of DNA denaturation at 95 °C for 5 min, followed by 32 cycles of 94 °C for 40 sec, 52-42 °C for 45 sec (the corresponding annealing temperature depending on each pair of primers, see Supplementary Table 1), and 72 °C for 40 sec, finally maintained at 72 °C for 5 min. Eight primers were selected for further analysis based on polymorphism and good band resolution. PCR products were supplemented with 5 (iL of loading buffer (40% w/v sucrose, 0.25% bromophenol blue, 0.25% w/v xylene cyanol). Electrophoretic separation of the PCR products was performed on 1% agarose gels at 70 V for 1 h in 1X TBE buffer with ethidium bromide (0.5 (ig/mL). The molecular profiles were visualized under UV light, photographed and stored for further analysis with the G-box (Syngene®).
Statistical analysis of the data
The analysis of 17 morphological variables was performed through analysis of variance (ANOVA). Mean separation tests were done using the Tukey test with the Bonferroni correction. Variables were transformed to meet normality and/or homoscedasticity as needed. Mean Euclidean Distance (MED) among individuals in each accession was calculated to estimate phenotypic variation within accessions. Principal component analysis (PCA) based on the standardized Euclidean distance was performed.
ISSR profiles were scored for presence (1) or absence (0) of homologous DNA bands. Genetic diversity was estimated in each accession by means of percentage of polymorphic loci (%P) and number of specific bands per accession. Dissimilarity between pairs of individuals was estimated through genetic distances (GD). Mean genetic distance (MGD) was calculated as the average of the genetic distances among individuals within accessions. Principal coordinates analysis (PCoA) was performed and individuals were plotted in a 2-dimensional graph representing the first 2 axes. All calculations were done using Infogen (Balzarini et al. 2010). Total genetic variation was partitioned via analysis of molecular variance (AMOVA). The statistical significance of the proportion of the total variance attributed to correlation between individuals within accessions relative to that of the total (PhiPT) was tested through 1,000 random permutations using the software GenAlEx (Excoffier et al. 1992; Peakall et al. 1995; Peakall and Smouse 2006).
Joint analysis
Both agro-morphological and molecular traits were used for the simultaneous characterization of accessions by means of a Generalized Procrustes Analysis (GPA) based on the components obtained from the PCA and PCoA analysis previously described (Bramardi et al. 2005).
Mantel's test t (Mantel 1967) was used to establish the relationship between both types of data. Statistical significance was determined using 1,000 random permutations. Analyses were conducted in R version 3.3.2, using package 'vegan'.
Results
Agro-morphological variation
Ten of the 17 agro-morphological variables studied here showed significant (P<0.05) differences among accessions (Table 3), while 3 of the remaining 7 showed differentiation at P<0.10 (data not shown). Within accessions, MED indicated that accession CU had the highest variability, EM presented the lowest distance among individuals, and the remaining 6 presented values close to the lowest one (Table 3).
Table 3 Mean (M), Bonferroni test (Bt), relative performance (Rp) and mean Euclidean distance (MED) in Panicum coloratum var. coloratum accessions for 17 agro-morphological traits.
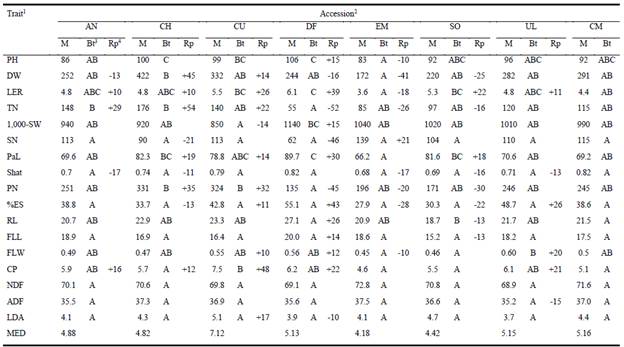
1Evaluated traits (codes, units): PH: Plant height (cm); DW: Dry weight per plant (g); LER: Leaf elongation rate (cm/tiller/d); TN: Number of tillers per plant; 1,000-SW: 1,000 seed weight (g); SN: Number of seeds per panicle (SN); PaL: Panicle length (cm); Shat: Seed retention; PN: Number of panicles per plant; %ES: Percentage of empty seeds; RL: Rachis length (cm); FLL: Flag leaf blade length (cm); FLW: Flag leaf blade width (cm); CP: Crude protein (g/100 g DM); ADF: Acid detergent fiber (g/100 g DM); NDF: Neutral detergent fiber (g/100 g DM), LDA: Lignin (g/100g DM). 2 Accession codes: AN: Anguil; CH: Chacharramendi; CU: Concepcion del Uruguay; DF: Dean Funes; EM: El Mirador; SO: Sol de Mayo; UL: Ultracán; CM: cv. Klein. 3 Bonferroni test: Different letters at each population indicate significant differences among populations at P<0.05. 4 Rp: Positive and negative numbers indicate higher or lower values than CM (cv. Klein), respectively.
Regarding accession characteristics, on average, CH plants showed the highest forage production and number of tillers, while EM presented the lowest dry weight per plant and DF the least number of tillers. However, differences in leaf elongation rate were not so pronounced, and populations presenting the highest values were DF, CU and SO. Forage quality showed a different pattern, with CU plants having highest crude protein concentration and EM plants the lowest, while differences between accessions in NDF, ADF and lignin concentrations were generally less than 10% (P>0.05). All materials produced similar numbers of seeds per panicle (P = 0.32) and showed only marginal differences in the percentage of empty seeds (P = 0.08).
A PCA biplot based on phenotypic variability among individuals explained 37.7% of the variation in the first 2 axes. Nine variables attained percentages of reconstruction approaching or exceeding 50%, while 3 variables showed very low reconstruction in the first 2 axes (Figure 1A) . In general, individuals did not congregate according to their origin.
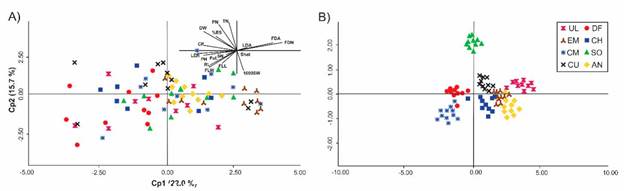
Figure 1 A) Individual and variable plots of principal component analysis (PCA) based on the individual distance matrix of Panicum coloratum var. coloratum calculated from 17 agro-morphological traits. B) Principal coordinates analysis (PCoA) plot based on the individual ISSR distance matrix of P. coloratum var. coloratum. See Tables 1 and 2 for a fuller description of the accessions and variables evaluated.
Comparing the relative performance of accessions with that of cv. Klein (CM), currently commercialized in Argentina, some accessions out-performed Klein in LER, PaL and CP, demonstrating that there is some room for selection.
Molecular variation
Analysis of 80 plants of P. coloratum var. coloratum rendered 127 ISSR reproducible bands, and 65 of them showed polymorphism (51.2%). The number of amplified bands per primer ranged from 10 to 24, representing a polymorphism range from 40.1 to 67.5% (data not shown).
The amount of variability among individuals within accessions did not differ between accessions: the percentage of polymorphic loci ranged from 42.5 in DF and SO to 59.1 in AN (Table 4). Cultivar Klein (CM) showed an intermediate level of variability. The same pattern was observed regarding mean genetic distance (MGD), with accession AN being the most variable, while SO displayed the least MGD between individuals. Accession-specific ISSR bands were apparent only in AN, SO and CM, indicating that most bands were shared by at least 2 accessions.
Table 4 Genetic diversity parameters for ISSR markers in accessions of Panicum coloratum var. coloratum.
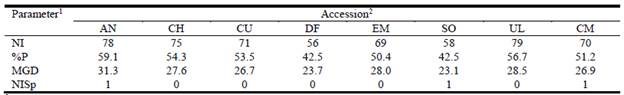
1NI: total number of loci; %P: percentage of polymorphic loci; MGD: mean genetic distance; and NISp: number of accession-specific loci. 2Accession codes see Table 1.
The first 2 axes of the PCoA analysis based on the ISSR distance matrix explained 20.0% of the variation (Figure IB) . Four groups could be identified in the plot, although the groups were spread and showed some level of overlapping between them. A rather distinct group was formed by individuals of SO, well separated from the rest. Individuals from AN, EM, CU and CH clustered in the center and formed a group. DF appeared related to CM, and UL showed dispersed individuals forming a separated group.
The dendrogram based on the individual GD ISSR distance matrix clearly conglomerated individuals from the same origin but at a high distance (Figure 2, r = 0.83). AMOVA performed on ISSR data showed that only 42% of the molecular variability is attributable to differences between accessions; therefore, most of the variation in this variety would be among individuals within accessions. In agreement with that, only 2 of the assayed primers showed more that 50% of the differentiation between accessions, while another 2 initiators indicated that more than 75% of the total genetic variation was explained by differences between individuals within accessions.
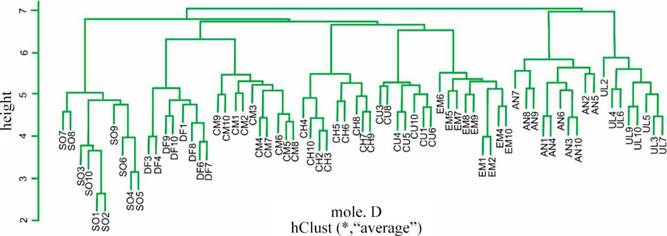
Figure 2 Unweighted pair-group method with arithmetic average (UPGMA) dendrogram based on the GD ISSR distance matrix of Panicum coloratum var. coloratum. See Table 1 for a fuller description of the accessions evaluated.
Joint characterization
From the Generalized Procrustes Analysis, the first 2 components were responsible for 20.5 and 16.3% of the total variation, respectively, adding up to 36.8%. Breaking down the total sum of squares showed that 77.3% corresponded with the consensus configuration, implying that only 22.7% was linked to the divergence between agronomic and molecular points and the points of the consensus configuration. In addition, the results of the Mantel test showed that the correlation between the 2 sets of data was very low (r = 0.057; P = 0.082).
Discussion
This study reported significant variation for the 17 morphological and molecular characters evaluated in the collection of P. coloratum var. coloratum at INTA in Argentina, a collection allotted to germplasm preservation and breeding. Even though the species has been an introduced forage to Argentina and introductions were not well documented, it can be argued that no reduction in variability due to factors such as, for example, founder effect occurred. The collection was assembled from ramets of adult plants, sometimes from quite old pastures that might have been under high selection pressure exerted by grazing, but no reduction of variability relative to the original populations was observed (Silvertown and Lovett Doust 1993). The most variable characters were related to forage production (number of tillers and dry weight per plant) and seed production (number of panicles per plant and number of seeds per panicle), as has been reported for other forages (Casler 2005; Morales-Nieto et al. 2008; Wójtowicz et al. 2009; Abbott and Pistorale 2010). As plants were grown in a common garden setting, environmental conditions were similar for all.
Variability in agro-morphological traits was found both within and between accessions that were in general quite variable, with a certain degree of overlapping. Although some accessions were significantly different for some characteristics, it may be difficult to allocate individuals to accessions based only on morphology. The same degree of variability observed within accessions was registered in the commercial material, CM (cv. Klein). Clearly seed of cv. Klein marketed in Argentina is from a rather genetically wide stabilized population of an allogamous species and identification of the origin of the seeds by a governmental agency may not be simple. This cultivar was developed in the USA (Carr 2014) through a process of recurrent selection to increase seed weight (Hussey and Holt 1986). In fact, the collection at INTA comprises accessions with heavier seeds, opening the possibility of a new cycle of selection. An important short-coming of the species is the difficulty in seed production due to shattering (Young 1994), which causes major losses of seed at harvesting. Improving this characteristic would be a worthwhile aim for a breeding program. While some materials with greater seed retention have been registered in the USA (Tischler and Ocumpaugh 2004), we are unaware of any attempts to improve this character through breeding programs. Since variability in this collection is located within accessions it is possible that, even though the commercial material and all accessions showed comparable levels of seed shattering, some individuals with higher seed retention may be identified for use as parents in future breeding programs.
In this study, there was some variability among populations in terms of CP concentration in the forage and accession CU actually produced forage of better quality than that from the commercial cultivar being marketed in the country. Although forage production does not necessarily predict secondary production (meat, milk or wool), forage with higher CP might be more digestible and give higher animal performance (Meissner 1997; Smith et al. 1997). Increments of 1% in in vitro digestibility of forage can lead to an increase in animal liveweight gain of 3.2% (Casler and Vogel 1999).
The high levels of variability within and among accessions of the collection as shown by the use of 8 ISSRs (Table 4) are similar to the genetic diversity observed in other species of open-pollinated grasses, e.g. zoysiagrass (Xie et al. 2012b) and bermudagrass (Cynodon dactylon) (Huang et al. 2010). Accession AN showed the highest level of variability, with approximately 59% of polymorphism, the highest MGD and 1 of the 3 accession-specific loci. Combining these features, AN emerged as a significant reservoir of variability in the collection.
As previously discussed, analysis using agro-morphological data failed to detect any type of association between accessions. In contrast, PCoA analysis showed some degree of grouping among accessions according to their site of collection as previously reported in the literature (García et al. 2007; Najaphy et al. 2012). In addition, the dendrogram obtained from the molecular data grouped individuals from the same origin, although the grouping was attained at a high distance, supporting the fact that 58% of the variation was found between individuals within groups, with only 42% between groups. A rather cohesive group was formed by accessions CM and DF, the latter being a recently sown pasture, so it could be hypothesized that individuals of DF retained characteristics of the commercial material, given that they were not exposed to grazing or environmental conditions for enough time to change their genetic composition. On the opposite side of the plot, UL could be identified as an accession from a rather old pasture that had probably been under heavy grazing pressure for several years. Accessions in the center of the plot aggregated into 2 groups: the one below showed populations CH, AN and EM in clusters; CH and EM had been recently sown from seeds produced at the AN site, so shared the same genetic material. A quite separate group was formed by individuals of accession SO obtained from imported seeds with a history of heavy grazing that might have increased the proportion of individuals with grazing tolerance. Finally, the history of CU is undocumented.
The AMOVA study indicated that more than 58% of molecular variation in P. coloratum resides within accessions. This finding is consistent with the predominant allogamous form of reproduction in the species (Hamrick and Godt 1990). Similar patterns of genetic variation distribution were reported in other cross-pollinated species. Xie et al. (2012a) reported 65% variation within accessions of orchardgrass (Dactylis glomerata) using SSR. Panicum coloratum var. coloratum is reported as having a high rate of sexual reproduction (Tischler and Ocumpaugh 2004), although the ability to reproduce asexually by apomixis should not be disregarded. In addition, vegetative reproduction through rhizomes and rooting at the nodes when they come into contact with the ground has been observed (Petruzzi et al. 2003; Komatsu et al. 2007).
Results of characterization using molecular markers and agro-morphological descriptors produced different configurations (Figures 1A and 1B). In addition, results of the Mantel test (r = 0.057; P = 0.082) confirmed low correlations between the two types of data, indicating that each individual characterization offers information that can be considered complementary. Semagn (2002) attributed the discrepancy between morphological and molecular variation to 2 possible factors: 1) molecular markers covering a large portion of the genome that includes coding and non-coding regions; and 2) molecular markers being less subject to artificial selection than morphological markers. According to Holderegger et al. (2006), most DNA markers are a random sample whose genome polymorphism sites have no phenotypic effects corresponding to neutral genetic variation, while quantitative morphological characters are usually affected by selection and variability within them is ultimately responsible for adaptation. Interestingly, joint characterizations rendered a consensus configuration explaining 77.3% of the variability, emphasizing the importance of studying the different types of descriptors together in order to get the best characterization and interpretation of the genetic diversity of the collection.
It is widely acknowledged that good characterization of a live collection is an important first step towards germplasm management and preservation. The collection of P. coloratum var. coloratum at INTA Rafaela was found to be a good resource and a reservoir of genetic variation for valuable characters to be further used in breeding for different objectives such as augmentation of forage production, forage quality or seed weight. The use of both molecular and agro-morphological traits was a valuable tool as both types of characterizations were complementary and jointly rendered a good discrimination of accessions.