Introduction
Chloris gayana Kunth, a gramineous perennial species native to Africa, is one of the most important warm-season forage grasses in subtropical and tropical areas of the world (Ponsens et al. 2010). Currently, this species is being introduced into temperate areas as a consequence of increasing minimum temperatures (i.e. less severe winters) because of global warming (Imaz et al. 2015), so its importance as a forage species is increasing worldwide. Its important characteristics are high forage production (up to 15 t DM/ha; see Bogdan 1963), seed production (up to 200 kg/ha; see Bogdan 1963) and some degree of tolerance of major abiotic stresses, such as cold (Cook et al. 2005), drought (Ponsens et al. 2010), flooding (Imaz et al. 2015) and soil salinity (Loch et al. 2004). Chloris gayana is a cross-pollinated species that includes diploid as well as tetraploid cytotypes, and the degree of salt tolerance and other agronomic attributes vary among populations or commercial cultivars (Loch et al. 2004).
Different techniques have been used to evaluate genetic variability. Primarily, morphological and agronomic attributes are employed for establishing genetic relationships and to discriminate among populations or commercial cultivars (Loch et al. 2004). More recently, molecular markers have been used for determining genetic variability among and within tetraploid and diploid cultivars/cytotypes (Ubi et al. 2000, 2003). Knowledge of genetic relationships among cultivars is an important consideration for classification and utilization of germplasm resources and breeding (Baretta et al. 2016). When different characterization systems are used, it is necessary to determine the degree of consistency between the different methods. One method used is Generalized Procrustes Analysis (GPA) (Jana et al. 2017). The use of combined datasets has been recommended because each dataset provides complementary information allowing greater power of resolution in genetic variation analysis, and this has proved to be superior to other methods for characterizing plant germplasm (Alves et al. 2013). However, a comparative analysis (consensus) using molecular and agronomic data has not been reported for Chloris gayana.
Salinization is one of the oldest and most severe environmental problems in the world, affecting more than 930 M ha in more than 100 countries (Bazihizina et al. 2012). Saline soils can limit establishment, persistence and forage production, especially for perennial pasture species, by affecting plant growth at various stages of development, including germination, seedling emergence and vegetative growth (Munns and Tester 2008). Ability to germinate is particularly important in perennial grasses with small seeds, e.g. Chloris gayana, for successful pasture establishment in saline soils (Quiroga et al. 2016). In addition, early vegetative growth in forage grasses is usually constrained under saline conditions (De Luca et al. 2001). Understanding plant responses to high salinity stress and subsequently selecting or developing salt-tolerant varieties can be a solution for increasing livestock production in marginal areas for agriculture, with restrictive soil-climatic conditions (Ashraf and Akram 2009).
Therefore, the objectives of this work were to: 1) characterize diploid unselected populations and synthetic varieties of Chloris gayana through agronomic traits and molecular markers; 2) establish the degree of consensus between groupings based on agronomic and molecular data; and 3) evaluate responses to salinity stress at germination and in early vegetative growth stages in the diploid synthetic varieties of C. gayana.
Materials and Methods
Agronomic characterization
The experiment was conducted in an experimental field located in Córdoba, Argentina (31º28' S, 64º14' W; 600 masl) on a loamy sand corresponding with an Entic Haplustoll soil, with 2.5% organic matter and pH 7.1. Five diploid C. gayana cultivars were tested: 2 unselected populations (Pioneer and Katambora) and 3 synthetic varieties (Topcut, Finecut and Santana). Topcut and Finecut are cultivars selected from Pioneer and Katambora, respectively, for hay production in Australia (Loch et al. 2004), whereas cv. Santana was obtained from C. gayana accessions derived from West Africa for salt tolerance in Argentina (Ribotta et al. 2013). All of these materials are used as commercial cultivars in Argentina.
Forty plants of each cultivar were transplanted to a field in a randomized complete-block design with 4 replications of 10 plants each. Distance between plants in a plot and between plots was 1 m. Twenty-five agronomic traits were measured (Table 1).
Molecular characterization
Total genomic DNA was extracted from leaves of each cultivar using the Nucleon PhytoPure Genomic DNA Extraction Kit (GE Healthcare Life Science, Amersham, UK). For Sequence-related Amplified Polymorphism (SRAP) markers, 20 primer pair combinations were tested; they were designed from previous work in other plant species (Li and Quiros 2001; Castonguay et al. 2010). Polymerase chain reactions (PCRs) were performed in volumes of 20 μL, using a protocol modified from Li and Quiros (2001). A total of 18 Inter Simple Sequence Repeats (ISSRs) primers were tested based on common primer selections reported for other Gramineae species (Gutiérrez-Ozuna et al. 2009; Li et al. 2011). The PCRs were performed in volumes of 20 μL, as described by Gutiérrez-Ozuna et al. (2009) with minor modifications. All PCR reactions were performed on an Eppendorf Mastercycler (Eppendorf AG, Hamburg, Germany). The amplifications were repeated twice and only clear repetitive bands were used in data analysis. Amplification products were loaded on a 2% agarose gel stained with ethidium bromide (10 mg/ml) and run for 3 h at 50 V. DNA fragments were visualized using a BIORAD, Molecular Imager® Gel DocTM XR System (USA). The SRAP and ISSR bands were scored as absent (0) or present (1) if visible, regardless of their relative intensity.
Statistical analysis
The agronomic data were subjected to analysis of variance (ANOVA) and the means were compared by the DGC test (Di Rienzo et al. 2002) at P≤0.05 using InfoStat software (Di Rienzo et al. 2016). In addition, cluster analysis was performed through the unweighted pair-group method using an arithmetic average (UPGMA) algorithm based on the standardized Euclidean distance.
For SRAP and ISSR markers, distance matrix was based on Jaccard coefficients [sqrt (1-S)], and a dendrogram was created using UPGMA. A cophenetic coefficient was computed from the clustering matrix to compare the matrix of genetic similarity and the dendrogram. The number of total (TB), monomorphic (MB) and polymorphic (PB) bands and polymorphic information content (PIC) were calculated for each primer or primer combination.
A Generalized Procrustes Analysis (GPA) was performed using the Gower distance (Gower 1975) to measure consensus between the agronomic and molecular information. A principal component (PC) analysis and Minimum Spanning Tree (MST) were performed on each of the distance matrices.
The above-mentioned analyses were performed using InfoStat (Di Rienzo et al. 2016) and InfoGen (Balzarini and Di Rienzo 2011) software.
Evaluation of salinity response at germination and early vegetative stages
The evaluations of salinity response during germination and early vegetative stages were performed in the synthetic varieties of C. gayana (Topcut, Finecut and Santana) due to these materials being the most divergent when agronomic and molecular characterization were realized (analysis consensus).
For the evaluation of salinity response at the germination stage, the assay was performed according to López Colomba et al. (2013) with minor modifications. In brief, seeds of these 3 cultivars were surface-sterilized for 20 min in 10% commercial bleach (NaClO 55 g/L), rinsed 3 times in sterile distilled water and transferred to plastic trays lined with filter paper soaked with each salt treatment. The salt treatments used were 0 mM (control), 100, 200 and 300 mM NaCl (approximately 0, 10, 20 and 30 dS/m, respectively). Seeds were incubated in a growth chamber (LIADE, Laboratorio de Investigación Aplicada y Desarrollo, Córdoba, Argentina) under the following conditions: 8 h light/16 h dark photoperiod (55 µmol/m2/s) at alternating temperatures of 25 ºC (8 h) and 20 ºC (16 h). Four replicates of 30 caryopses per treatment and per cultivar were used. Germinated seeds were counted every 72 h for 21 days, discarded after counting and the proportion of germinated seeds was calculated.
To evaluate salinity response during early vegetative growth, the assay was conducted in a greenhouse under hydroponic conditions. Seedlings with 4 leaves developed were placed individually in holes of a Styrofoam board. These boards were set on rectangular plastic trays (30 × 20 × 60 cm) filled with aerated Hoagland nutrient solution (Hoagland and Arnon 1950). Eighty seedlings of each cultivar were randomly located in 4 trays, with 20 seedlings per tray; each tray was treated as an experimental unit. Two trays were allocated to the control (nutrient solution without NaCl) and the two other trays, to the saline treatment of 400 mM NaCl. Salinization was accomplished by gradually adding 100 mM NaCl every 24 h to avoid osmotic shock, until a concentration of 400 mM NaCl (approximately 40 dS/m) was reached. After 30 days of treatment, the seedlings were evaluated for the following traits: aerial, root and total fresh weight (AFW, RFW and TFW, respectively); aerial, root and total dry weight (ADW, RDW and TDW, respectively); root length (RL); number of stems (SN); and plant height (PH). The seedlings were dried at 60 ºC in a forced air oven for 48 h to achieve stable weight.
Statistical analysis
Effects of cultivar, salt concentration and their interactions on the salt tolerance at germination and early vegetative growth stages were determined by Generalized Linear Mixed Models to estimate differences among cultivars and salt concentrations. The cultivars, salt concentrations and their interactions were considered fixed effects, whereas each repetition was considered a random effect. The average values were compared using Fisher´s LSD test. The above-mentioned analyses were performed using InfoStat (Di Rienzo et al. 2016) software.
Results
Agronomic and molecular characterization
Twenty-two agronomic traits revealed differences among cultivars (Table 2) and the most discriminating traits were those related to morphological and reproductive aspects.
Table 2 Means of 4 replicates (± s.d.) of agronomic traits measured in diploid unselected populations and synthetic varieties of Chloris gayana.
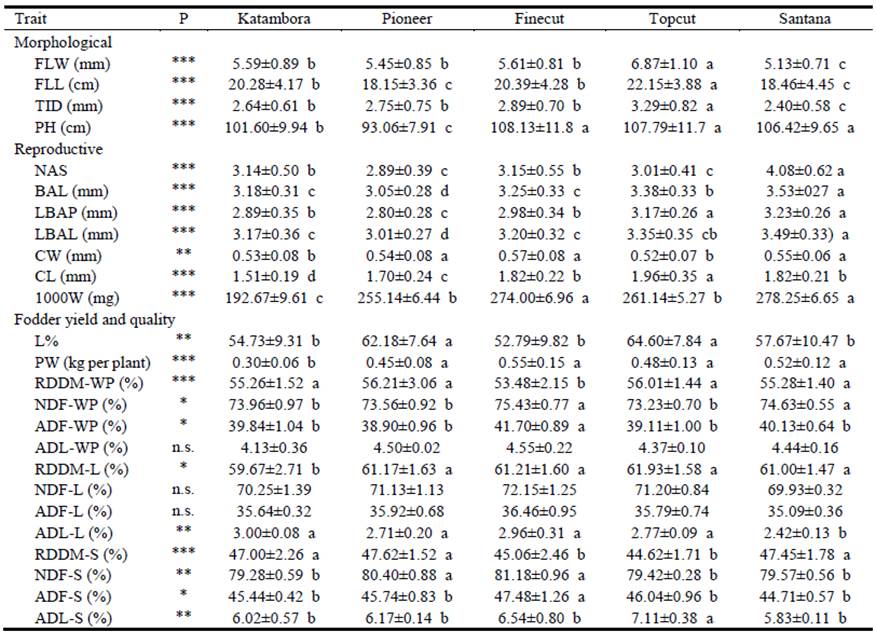
Abbreviations: FLW (Flag leaf width), FLL (Flag leaf length), TID (Tiller diameter), PH (Plant height), NAS (Number of anthecia), BAL (Basal anthecium length), LBAP (Length of basal anthecium palea), LBAL (Length of basal anthecium lemma), CW (Caryopsis width), CL (Caryopsis length), 1000W (Weight of 1,000 caryopses), L% (leaf percentage), PW (Plant weight), RDDM (Ruminal disappearance of dry matter), NDF (Neutral detergent fiber), ADF (Acid detergent fiber), ADL (Acid detergent lignin). For RDDM, NFD, ADF and ADL, companion letters mean WP (whole plant), L (leaves: leaf blades plus sheaths) and S (stems plus inflorescences).
Flag leaf width, flag leaf length and tiller diameter were greatest in Topcut and least in Santana (Table 2). Regarding plant height, the unselected populations (Pioneer and Katambora) were shorter than the synthetic varieties (Topcut, Finecut and Santana; Table 2). In general, Santana stood out above the remainder in terms of reproductive characters. It displayed greater numbers of anthecia, basal anthecium length and length of basal anthecium lemma than most other cultivars, whereas Pioneer showed the lowest values for the BAL, LBAP and LBAL. Caryopsis width, caryopsis length and weight of 1000 caryopses also demonstrated differences between cultivars (Table 2).
In relation to fodder yield, Katambora showed significantly lower plant dry matter (DM) than the remaining 4 cultivars (Table 2). The important indicator of potential feeding value, leaf percentage, was significantly higher in Pioneer and Topcut (mean 63.4%) than in the other 3 cultivars (mean 55.1%). In relation to ruminal disappearance of DM, the disappearance of whole plant material for Finecut (53.5%) was lower than for the remainder (mean 57.6%), while disappearance of leaf was less in Katambora (59.7%) than in the other cultivars (mean 61.4%) (Table 2). Disappearance of stem was lower in Topcut and Finecut (mean 44.8%) than in Santana, Katambora and Pioneer (mean 47.4%). Finecut had the highest percentages of acid and neutral detergent fiber in whole plant and stems. Topcut showed the highest values for acid detergent lignin in stems, while Santana showed the lowest value in leaves (Table 2).
As for molecular characterization, 20 SRAP primer combinations showing reproducible and polymorphic patterns generated a total of 256 scorable bands with an average of 12.8 bands per primer combination, of which 82 were polymorphic (32.0%). The total number of bands produced by each primer combination ranged from 4 (F9/R8) to 22 (F13/R15), while the number of polymorphic bands produced by each primer combination ranged from 1 (ME2/R9) to 8 (F13/R15 and ME4/R8) (Table 3); ME2/EM2 and ME2/R9 were the primer combinations with more polymorphic information content (PIC) (0.36). It is noteworthy that Finecut, Santana and Topcut showed numerous unique markers (17, 8 and 3, respectively), whereas Pioneer and Katambora showed none. Eighteen ISSR primers amplified a total of 306 bands, of which 147 were polymorphic (48.0%) with an average of 17 bands per primer. The number of polymorphic bands ranged from 4 (UBC864, UBC862 and UBC826) to 15 (SC ISSR1); UBC864 and D12 ISSR had the highest PIC (0.36) (Table 3). Cultivars Finecut, Topcut and Santana showed numerous unique markers (25, 14 and 8, respectively), which was similar to the pattern observed for SRAP markers.
Table 3 Number of polymorphic bands (PB), monomorphic bands (MB), total bands (TB) and polymorphic information content (PIC) revealed by 20 SRAP primer combinations and 18 ISSR primers in diploid unselected populations and synthetic varieties of Chloris gayana.
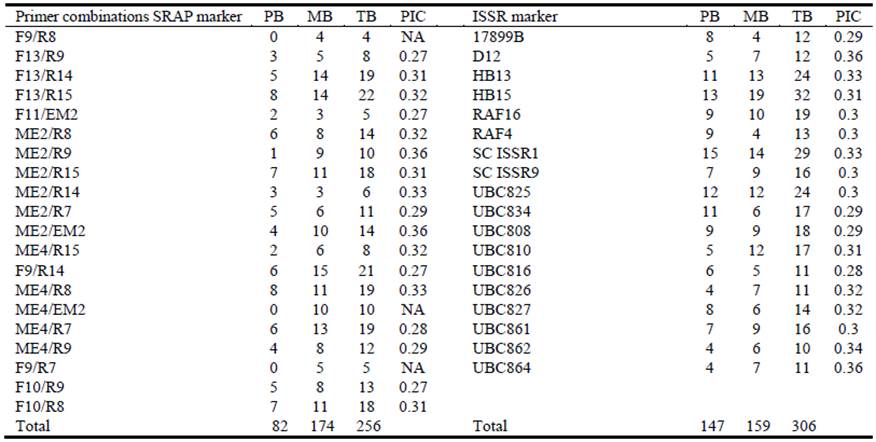
NA: Not available.
A dendrogram based on UPGMA analyses of the 25 agronomic traits revealed that the 5 cultivars constituted 2 major clusters (Figure 1a; genetic similarity coefficient range 4.65‒7.70; cophenetic correlation coefficient = 0.97). Cultivar Finecut, which formed a separate cluster, was genetically different from the remaining cultivars, which formed a second group. While Pioneer and Katambora were closely related, Santana was intermediate between them and Topcut (Figure 1a). The dendrogram based on UPGMA analyses of the SRAP (genotypic) data showed a similar trend to that obtained with agronomic analysis (Figure 1b; genetic diversity coefficient range 0.24‒0.46; cophenetic correlation coefficient = 0.95). Finally, the ISSR dendrogram exhibited a trend to form clusters similar to those based on agronomic traits and SRAP markers, with the exception of Topcut, which showed close relationship with Pioneer and Katambora (Figure 1c; genetic diversity coefficient range 0.33‒0.59; cophenetic correlation coefficient = 0.99).
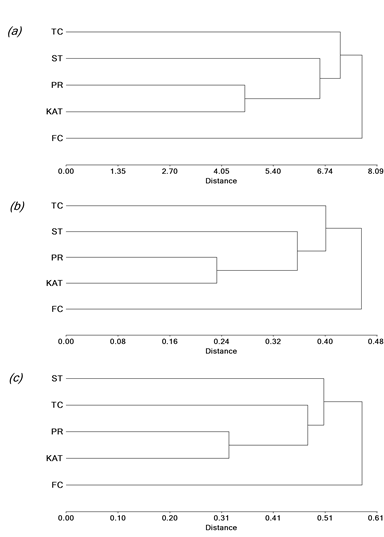
Figure 1 UPGMA dendrogram showing genetic relationships among diploid unselected populations and synthetic varieties of Chloris gayana. The relationships were calculated on the basis of genetic distances using: a) 25 agronomic traits; b) 20 SRAP markers; and c) 18 ISSR markers. Katambora (KAT), Pioneer (PR), Finecut (FC), Topcut (TC) and Santana (ST).
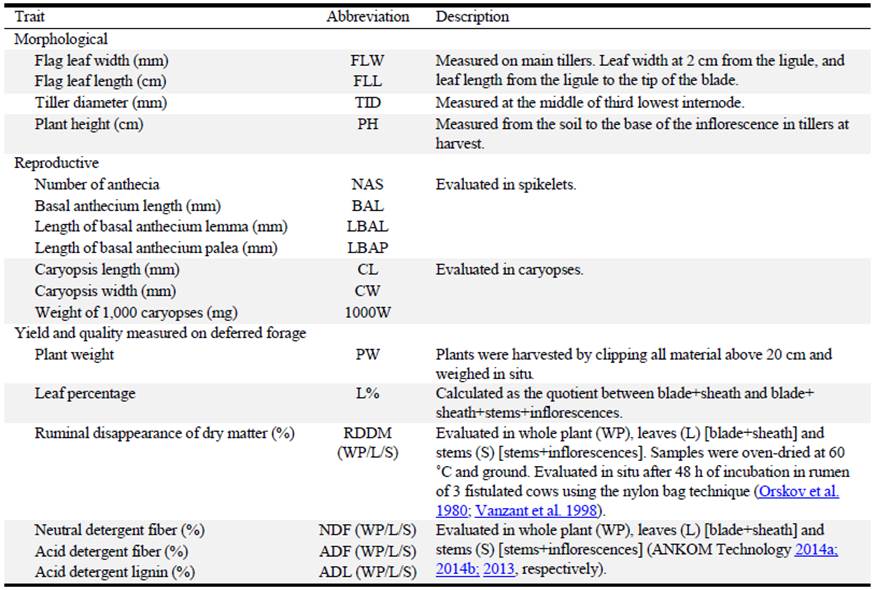
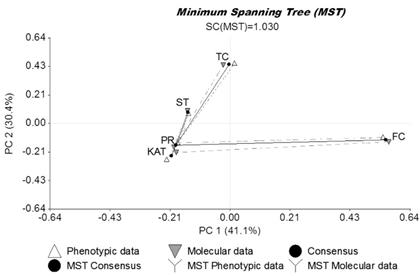
Generalized Procrustes Analysis showed that: 1) 71.5% of the total variability contained in the datasets was explained by the first 2 axes (PC1: 41.1% and PC2: 30.4%); and 2) there was high correspondence between agronomic and molecular datasets (99.9% consensus) (Figure 2). Cultivars Finecut and Topcut were distinct and different from other cultivars whereas the unselected populations, Pioneer and Katambora, appeared to be closely related. Cultivar Santana showed some relationship with Pioneer and Katambora (Figure 2).
Evaluation of salinity responses during germination and early vegetative stages
The cultivars responded differentially to salinity in terms of germination percentage with significant cultivar × salt concentration interactions (P = 0.035; Table 4). All cultivars recorded excellent germination rates (mean 96%) in water, and cultivars Finecut and Santana still displayed high values in saline conditions (means of 96 and 93% for 100 mM and 200 mM NaCl solutions, respectively; Table 4). By contrast, germination of cv. Topcut declined to 82% in both 100 and 200 mM NaCl solutions. At the highest saline concentration (300 mM NaCl), all cultivars exhibited reductions in seed germination relative to control (87% for Finecut, 76% for Santana and 72% for Topcut; Table 4).
Table 4 Proportion of germinated seeds of diploid synthetic varieties of Chloris gayana at increasing salinity levels.
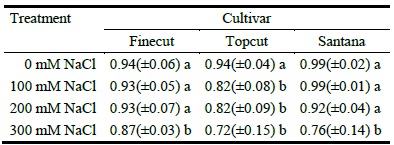
Mean values of 4 replicates (± s.d.) are presented. Different letters indicate significant differences (P<0.05) by LSD Fisher´s test.
In terms of early vegetative growth, cultivars responded differentially to 400 mM NaCl in aerial dry weight (ADW), root dry weight (RDW), total dry weight (TDW), maximum root length (MRL) and plant height (PH) (P<0.05; Table 5). For all parameters other than ADW, the interaction cultivar × treatment was significant (P<0.05). Under stressful saline conditions, Finecut showed less depression in growth than Santana and Topcut. ADW of Finecut in saline solution was 55.1% of that in straight water, while corresponding values for Santana and Topcut were 51.4 and 44.8%, respectively. Similarly, RDW for Finecut in saline solution was 80% of that in water, while values for Santana and Topcut were 56 and 52%. This resulted in TDW values in saline solution relative to straight water for Finecut, Santana and Topcut of 58.8, 50.4 and 46.9%, respectively. Despite these outcomes, in absolute terms Santana produced more TDW in saline solution than Finecut and Topcut. Topcut was the most sensitive in reaction to salinity, showing the largest reductions in aerial dry weight, root dry weight, total dry weight, plant height and maximum root length (Table 5).
Table 5 Plant growth parameters for 30 days after germination of diploid synthetic varieties of Chloris gayana under non-saline (0 mM NaCl) and saline (400 mM NaCl) conditions in hydroponic culture. Means of 2 replicates (40 plants) (± S.D.) for aerial dry weight (ADW), root dry weight (RDW), total dry weight (TDW), number of stems (SN), plant height (PH) and maximum root length (MRL).
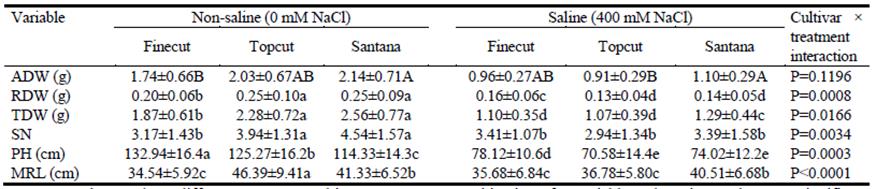
Lower-case letters show differences among cultivar × treatment combinations for variables where interaction was significant (P<0.05). Uppercase letters show significant differences among cultivars within treatments for variables where interaction was not significant (P>0.05).
Discussion
This study presents 3 major contributions: 1) the characterization of diploid unselected populations and synthetic varieties of C. gayana via agronomic traits and SRAP and ISSR molecular markers; 2) the identification of high consensus (>99%) between molecular markers and agronomic traits, suggesting that both systems provided similar estimates of genetic relationships; and 3) novel evidence showing existence of variability in salinity tolerance among diploid synthetic varieties of C. gayana.
Agronomic characterization allowed us to differentiate between the evaluated cultivars. While Agnusdei et al. (2009) suggest there is a negative association between leaf dimensions and nutritive quality, cv. Topcut showed the highest values for traits related to leaf dimensions in our study without detriment to its nutritional value. In a study conducted in Cenchrus spp. by Griffa et al. (2012), the morphological traits width and length of flag leaf and 1,000-seed weight were closely related to seed production. In fact, seed production was negatively correlated with leaf size but positively correlated with 1,000-seed weight. Our data support this relationship as cv. Santana exhibits low values for flag leaf length and width, but high values for seed-related characters. This relationship was confirmed by Cicetti et al. (2015) in a comparative study with other C. gayana diploid cultivars. The good performance of seed-related traits (weight, width and length) observed in Santana and Finecut suggests their potential for producing vigorous seedlings and enhanced performance at early stages of pasture establishment (Giordano et al. 2013). The results for fiber percentage and ruminal disappearance of dry matter were within the range of values reported for deferred C. gayana (Otondo 2004; Borrajo et al. 2015). These observations have significance for deferred forage utilization (Gargano et al. 2001).
In recent years, ISSR and SRAP markers have been recognized as low-cost useful molecular techniques in marker-assisted selection, for genetic linkage map construction and genetic diversity analysis (Castonguay et al. 2010; Shao et al. 2010). Our results suggest that ISSRs were superior to the SRAP markers in their capacity to produce polymorphic bands, which is in agreement with previous reports for Chrysanthemum morifolium (Shao et al. 2010), Cynodon arcuatus (Huang et al. 2013) and Prunus armeniaca (Li et al. 2014). The differences between the methods might be related to the fact that, unlike ISSRs, which are targeted to micro-satellite regions in the whole genome, SRAP markers preferentially amplify open reading frames targeting functional regions of the genome (see Li and Quiros 2001).
It was interesting that clustering obtained from molecular and agronomic data revealed a grouping where the most closely related were the unselected populations, while the most genetically different cultivars were those obtained from breeding programs (Finecut, Topcut and Santana). The relationships between cultivars Pioneer and Katambora revealed by the analysis of ISSR and SRAP techniques are consistent with results reported by Pérez et al. (1999) using RAPD markers.
The information obtained by Generalized Procrustes Analysis (GPA) allowed us to establish a high consensus between the different data sets (agronomic and molecular) and suggested that both systems provided similar estimates of genetic relationships. Similar results were described by Bermejo et al. (2010) in the evaluation of recombinant inbred lines of lentils. In our study, the high degree of consensus obtained is probably a function of the high number of traits and molecular markers used. Even though GPA proved to be a very efficient tool to show the relationships among relative ordinations of a single genotype under different types of markers, it does not mean that it can replace/substitute for the information provided by each. The knowledge of quantitative and molecular variability can optimize the work of plant breeders (Ribotta 2011).
Seed germination is a critical phase when considering pasture establishment. Salinity imposes stresses on seeds during the germination process as soil water availability is reduced due to the low osmotic potential. Therefore, imbibition and germination of seeds can be constrained at increasing soil salinity (Munns and Tester 2008; Quiroga et al. 2016). Significantly, in this study differences were found among synthetic varieties under saline conditions. Finecut and Santana were able to germinate in the same proportion as controls even at concentrations of 200 mM NaCl, while germination in Topcut was suppressed at 100 mM NaCl. Curiously, Finecut and Santana have heavier seeds than Topcut, which might provide higher amounts of reserves to sustain germination under stressful conditions (Hanley et al. 2007). Grieve and Francois (1992) indicated that larger seeds frequently confer distinct advantages, e.g. seedling vigor and hardiness, to a species in overcoming salinity.
Germination and seedling establishment are considered the most critical stages of the plant life cycle under saline conditions. Salt tolerance in these ontogenic stages would confer distinct advantages to a species in terms of persistence and good performance in saline environments (Bazzigalupi et al. 2008). In a study conducted in Chloris gayana, salt tolerance conferred higher DM production (Moore et al. 2004), and provided higher numbers of tillers (De Luca et al. 2001). In our work, Santana showed better performance in total dry weight than Topcut or Finecut, whereas Finecut was the only cultivar that did not reduce its number of stems in saline conditions. These characteristics would allow survival of Santana and Finecut cultivars under saline conditions in the field. The root system plays a fundamental role in plant growth and survival because of water and nutrient uptake (Wang et al. 2009). It is known that plants will allocate relatively more biomass to roots if the limiting factor for growth is below ground (e.g. nutrients, water) (Poorter et al. 2012). In this work, Santana showed the longest roots, which would facilitate exploitation of mobile nutrients. According to Zolla et al. (2010), this characteristic should allow plants to compete better for resources under suboptimal soil conditions. Further analyses may be necessary to give insights to the mechanisms involved in the better response of Santana to salt stress under hydroponic conditions.
Summarizing, this study provides valuable information about agronomic and molecular characterization, which enables us to detect the existence of differences between unselected populations and synthetic varieties of C. gayana. Moreover, the agronomic and molecular characterization showed high consensus, suggesting similar estimates of the variability among cultivars. Our assessment of the 3 synthetic varieties in terms of their likely usefulness in saline environments suggests that Finecut and Santana show most promise for inclusion in forage planning exercises, given their high salinity tolerance at germination and early vegetative growth stages, both crucial phases when considering successful pasture establishment. Further field studies will be necessary to verify the hydroponic glasshouse results obtained in this study.