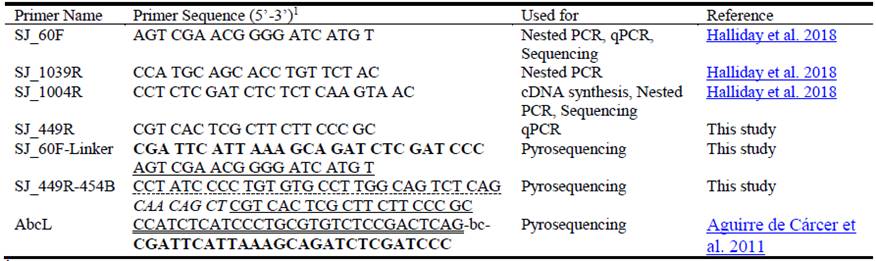
Introduction
Leucaena leucocephala is a nutritionally rich forage tree legume that contains a non-protein amino acid, mimosine, which is degraded by ruminal bacteria to the metabolites 3-hydroxy-4(1H)-pyridone (3,4-DHP) and 3-hydroxy-2(1H)-pyridone (2,3-DHP). Both these isomers of DHP are toxic and can result in impaired thyroid-like symptoms, chelation of metal ions, reduced liveweight gain and loss of appetite in animals. Raymond Jones discovered a bacterium in the rumen of Hawaiian goats that degraded these DHP metabolites into non-toxic end products (Jones and Lowry 1984), which was later isolated and named Synergistes jonesii (Allison et al. 1992). Subsequently, an inoculum containing S. jonesii was developed in Australia from a mixed rumen sample taken from a Hawaiian goat and has been used for many years as an ‘oral cattle drench’ in northern Australia (Jones and Meggarity 1986; Jones 1994; Klieve et al. 2002).
A survey of cattle herds grazing on leucaena in northern Australia showed high levels of 3,4- and 2,3-DHP excretion in urine, despite the majority of herds having been exposed to the ‘oral drench’ containing S. jonesii (Dalzell et al. 2012). The study did not attempt to detect S. jonesii, but concluded that a high level of 2,3-DHP (microbial metabolite of 3,4-DHP) in urine indicated its presence, albeit with incomplete degradation of the toxin (Dalzell et al. 2012). However other yet-to-be-discovered species of rumen bacteria, which degrade DHP isomers, could also be present in the rumen. Recently a new hypothesis has emerged which suggests that hepatic conjugation of DHP may contribute significantly to the detoxification of the isomers that have not undergone complete microbial degradation in the rumen (Shelton et al. 2019).
Molecular detection of the S. jonesii type strain (78-1, ATCC 49833) was first demonstrated by McSweeney et al. (1993) using radiolabelled (32P) and fluorescent-dye conjugated 16S rRNA targeted oligonucleotide probes. Subsequently, primer sets for PCR-based detection and enumeration of S. jonesii targeting genomic regions was developed by Yang et al. (1999) and further assessed for sensitivity by Anderson et al. (2004). However the nature/function of this template DNA in the S. jonesii genome was unknown, which raises doubts about the potential specificity of this detection method. Klieve et al. (2002) used a pair of 16S rRNA gene (rDNA) primers consisting of a universal bacterial primer (Primer 357F, Lane 1991) and DHP 1006 primer (McSweeney et al. 1993) to amplify a 438 nucleotide product specific for S. jonesii. Another set of 16S rDNA primers for S. jonesii (sng796f and sng1001r) were used by Derakhshani et al. (2015) but they did not report on their validation. In an attempt to increase sensitivity and specificity of detection, Graham et al. (2013) used a 16S rDNA nested PCR approach to monitor the presence of S. jonesii in cattle from northern Australian properties. This method detected S. jonesii in <10% of the cattle tested, even though several herds had been inoculated with the bacterium and DHP degradation was occurring. Sequence analysis of the 16S rDNA amplicons from samples positive for S. jonesii showed that all had differing sequence profiles compared with the S. jonesii ATCC type strain 78-1. Another survey of ruminants in different geographical regions confirmed the presence of S. jonesii in cattle, goats, sheep, yak and buffalo from Australia, China, Brazil, Thailand, Indonesia and Vietnam by an improved nested 16S rDNA PCR approach (Padmanabha et al. 2014). Sequence analysis of these PCR products revealed at least 4 loci with point mutations (single nucleotide polymorphisms; SNPs) compared with the ATCC type strain. The specificity of the PCR assay was further improved by Halliday et al. (2018) but showed <50% of rumen samples were positive for S. jonesii in a group of cattle partially degrading DHP. These studies indicate the bacterium is often present but below the limit of detection (104‒105 cells/mL) and therefore improved molecular assays are required for monitoring populations of S. jonesii in vivo.
The present study reports on further improvements in sensitivity of the PCR method for detecting the presence of S. jonesii in the rumen by using cDNA generated from rRNA as template for the PCR assay rather than its genomic DNA (gDNA). New primer sets were designed to amplify SNP regions of the 16S rDNA and these primers were also used in nested PCR, quantitative PCR (qPCR) and reverse transcriptase qPCR (RT-qPCR). This study also examined the geographical distribution relating to variations in the 16S rDNA of the S. jonesii strain 78-1 which may suggest divergence from the type strain in Australian cattle as well as in ruminants internationally. These changes may be correlated with the ability of the bacterium to degrade DHP, relative to the type strain.
Materials and Methods
Rumen fluid collection
Rumen fluid (RF) was collected mainly by orogastric tube from Australian cattle and from cattle, sheep, goats, buffalo, native cattle and yak from Indonesia, Thailand, Vietnam, China, Scotland and Brazil as well as gut samples or feces from some non-ruminant herbivores (see Table 2). Australian samples were generally stored on ice before freezing at -80 oC. Rumen samples from overseas were preserved in 100% ethanol for transportation at room-temperature prior to DNA extraction. Some rumen samples were also preserved for RNA extraction by mixing rumen fluid with an equal volume of RNALater TM Stabilization Solution (Thermo Fisher Scientific, Australia). These samples were chilled at ~2‒4 oC for 24‒48 h, centrifuged at 6,000 × g for 15 minutes and the pellet alone re-suspended in 70% ethanol for storage and trans-port at ambient temperature. Samples were stored at -80 oC until DNA or RNA extraction.
DNA extraction
Genomic DNA was extracted from 2 mL culture/rumen fluid pellets/feces, harvested by centrifugation for 20 min at 17,000 × g at 4 oC, using the cetyltrimethylammonium bromide (CTAB) method of Jones and Walker (1963) and Murray and Thompson (1980) with a modified bead-beating method (Gagen et al. 2010). Briefly, cell pellet from 2 mL rumen fluid sample was re-suspended in 800 µL of CTAB isolation buffer (100 mM Tris-HCl, pH 8; 1.4 M NaCl; 20 mM EDTA-disodium salt; 2% CTAB) and homogenized in a FastPrep®-24 bead-beater with zirconium-silica beads before extracting the DNA, quantifying and storing at -20 oC until used for PCR/qPCR analyses as described by Halliday et al. (2018). The gDNA from rumen digesta samples was diluted approximately to 50-100 ng/μL and used in a 25 μL PCR or qPCR.
RNA extraction and cDNA synthesis
Total RNA was extracted from samples either stored in RNALater TM or RNA Later TM followed by ethanol preservation. Synergistes jonesii pure culture cells harvested as standards for Reverse-Transcriptase qPCR (RT-qPCR) were initially ‘fixed’ by adding 1:1 volume of 5% phenol: ethanol (v/v), pH 4.3, mixed at 4 oC to prevent RNA degradation and the pellets stored frozen (-80 oC). A pellet from a 1 mL sample of culture or rumen fluid was harvested by centrifugation at 17,000 × g, 4 oC for 10 min and was re-suspended in (in order): 300 µL 10 × TE buffer pH 6.0 (Tris-EDTA buffer); 400 µL phenol:chloroform (1:1), pH 4.3, (Sigma-Aldrich, USA); and 100 µL 10% sodium dodecyl sulphate (SDS). Further, UV-sterilized 250 mg of zirconia/silica beads (0.1 mm and 1 mm, 1:1 w/w) was added to each tube and homogenized in a FastPrep-24® bead-beater (MP Biomedicals, USA) for 3 × 1 min at setting 6.5, with a rest of 1 min between cycles. The homogenized suspension was centrifuged as before and the aqueous phase transferred to a new tube for silica-gel column purification. The RNA was extracted using RNeasy® Mini Kit (QIAGEN GmbH, Germany) following manufacturer’s protocol with these modifications: 500 µl RLT buffer was added to the aqueous phase, mixed well, followed by adding 500 µL 96% ethanol to precipitate the RNA. This mixture was applied to an RNeasy Mini spin column, in 2 lots of 700 µL each and centrifuged at 8,000 × g for 15 sec to bind the RNA to the silica column and washed with RW1 wash buffer. Residual DNA contamination of the RNA was removed by on-column digestion using manufacturer’s DNase mix in RDD buffer and procedure, washed with RW1 wash buffer and twice with RPE buffer. The column was dried by a final centrifugation for 2 min at 10,000 × g and RNA eluted in 50 µL RNase free water and mixed with 1 µL RNaseOUTTM RNase Inhibitor (Thermo Fisher Scientific, Australia). The rRNA was quantified and RNA Integrity (RIN) assessed on an Agilent 2100 Bioanalyzer using Agilent RNA 6000 Nano chip Kit (Agilent Technologies, Santa Clara, CA) following the manufacturer’s instructions for prokaryotes. Total RNA was frozen at -80 oC.
Total RNA was converted to cDNA using the Superscript® III Reverse Transcriptase (Thermo Fisher Scientific, Australia) using 2 pmol of S. jonesii-specific primer (Sj_1004R: 5’-CCT CTC GAT CTC TCT CAA GTA AC-3’) following manufacturer’s protocol. Briefly, 1 µL of 2 µM S. jonesii-specific primer, 1 µL 10 mM dNTP mix and ~100 ng total RNA (typically 2 µL) was brought to a final volume of 14 µL and denatured at 65 oC for 5 min in a PCR machine and placed on ice for 3 min. To this, a master mix containing 4 µL, 5 × First-Strand buffer; 1 µL, 0.1 M DTT; and 1 µL, Superscript III RT (200 units/µL) was added and cycled in a PCR at 25 oC for 5 min; 50 oC for 60 min; and 70 oC for 15 min. The cDNA was frozen (-80 oC) or stored at 4 oC for analysis.
Detection of Synergistes jonesii by nested PCR and RT-qPCR
A nested PCR approach based on the 16S rDNA was used to detect the presence of S. jonesii using specific primer sets including new primers described by Halliday et al. (2018) (Table 1). The S. jonesii-specific cDNA generated from 16S rRNA was used as a template for RT-qPCR with gene-specific primers, Sj_60F and Sj_449R (Table 1), using SensiFAST™ SYBR® Lo-ROX Kit (Bioline Reagents Ltd, UK) following the manufacturer’s procedure. Each qPCR reaction was performed in quadruplicate (10 µL each) from a master mix with (1 µL/25 µL) cDNA template and aliquoted into MicroAmp® Optical 384-well reaction plate (Applied Biosystems, Life Technologies). The qPCR was run on Applied Biosystems ViiA™ 7 Real-Time PCR system with the following parameters: 95 oC for 3 min; 40 cycles of 95 oC for 15 sec and 60 °C for 1 min; and a final melt curve analysis consisting of 95 oC for 15 sec, 60 oC for 60 sec and 95 oC for 15 sec. Analysis was completed using QuantStudio™ Real-Time PCR software (Applied Biosystems, Carlsbad, CA). The sensitivity of detection of the nested PCR using gDNA compared with rRNA (converted to cDNA) was assessed by serial dilutions of mixed rumen culture seeded with S. jonesii 78-1 cells as described previously (Halliday et al. 2018). Amplicons from S. jonesii nested PCR-positive samples were Sanger sequenced to confirm identity as described in Halliday et al. (2018).
Survey S. jonesii SNPs using high-throughput sequencing
A survey of livestock from different countries for S. jonesii SNP distribution was done by 16S rDNA-based methods using a nested 454 pyrosequencing (454)-barcoding PCR as described by Aguirre de Cárcer et al. (2011) with minor modifications. Firstly, a primary PCR using S. jonesii-specific primer pair SJ_60F-SJ_1004R (Table 1) for 25 cycles was run. An aliquot (~5 µL) of each of this PCR product was cleaned using ExoI/CIAP (37 oC for 20 min and 80 oC for 20 min) followed by a 20 cycle nested PCR using primer pair SJ_60F-Linker-SJ_449-454B (Table 1) to generate S. jonesii 16S rDNA products (~380 bp V2/V3 region) for the SNP region. Finally, a 454-barcoding PCR run was completed on the cleaned 15‒20 µL aliquot of the positive nested-PCR products using unique 8 bp error-correcting barcodes with the 454 adaptor A and SJ_449R_454B primer pair (Table 1) for 10 cycles. The barcoded products were quantified using the Quant-IT dsDNA Assay kit (Thermo Fischer Scientific, Australia), pooled at equimolar amounts, concentrated, electrophoresed and gelpurified (QIAquick Gel Extraction Kit, QIAGEN GmbH, Germany). The final purified product was sequenced using a Roche 454 GS-FLX Titanium sequencer at Macrogen Inc. (Seoul, Korea).
Data analysis
Short read sequence data generated using 454 pyrosequencing for S. jonesii SNP patterns was analyzed using QIIME (Quantitative Insights Into Microbial Ecology) software package (Caporaso et al. 2010). Briefly, the sequences were demultiplexed based on barcode sequences and filtered for a minimal average quality score of 25 across a 50 bp sliding window and trimmed for length ranging from 300 to 600 bp. Raw sequences were passed through Acacia for 454-error correcting (Bragg et al. 2012). Error-corrected sequences were then clustered to OTUs at 100% similarity for S. jonesii performed using UCLUST algorithm (Edgar 2010). Chimeric sequences were identified using Chimera Slayer (Hess et al. 2011) and removed. Representative sequences were aligned to the Green Genes reference database (McDonald et al. 2012) using the RDP classifier software (Wang et al. 2007; Werner et al. 2012). Additional analysis of OTUs was performed in the R packages ade4 and Phyloseq (Chessel et al. 2004; McMurdie and Holmes 2013) and identification of the SNP pattern was achieved by filtering the alignment with a S. jonesii-specific lane mask for the 3 SNP-base positions.
Results
Detection of S. jonesii by nested PCR
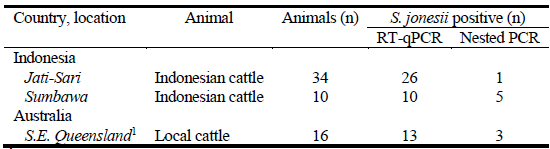
A two-step nested PCR approach was used to detect the presence of S. jonesii in gDNA extracted from gut contents of livestock in different countries. The primer pairs SJ_60F-SJ_1039R or SJ_60F-SJ_1004R were used in the primary PCR followed by SJ_60F-SJ_1004R or SJ_60F-SJ_449R primer sets (Table 1). Nearly all groups of animals tested from Australia, Indonesia, Thailand, Vietnam, Scotland, China and Brazil were positive for S. jonesii but rarely did all animals in a group return a positive result and <50% of the total tested positive (Table 2).
Table 2 Detection of S. jonesii in livestock from different countries using a nested PCR assay.
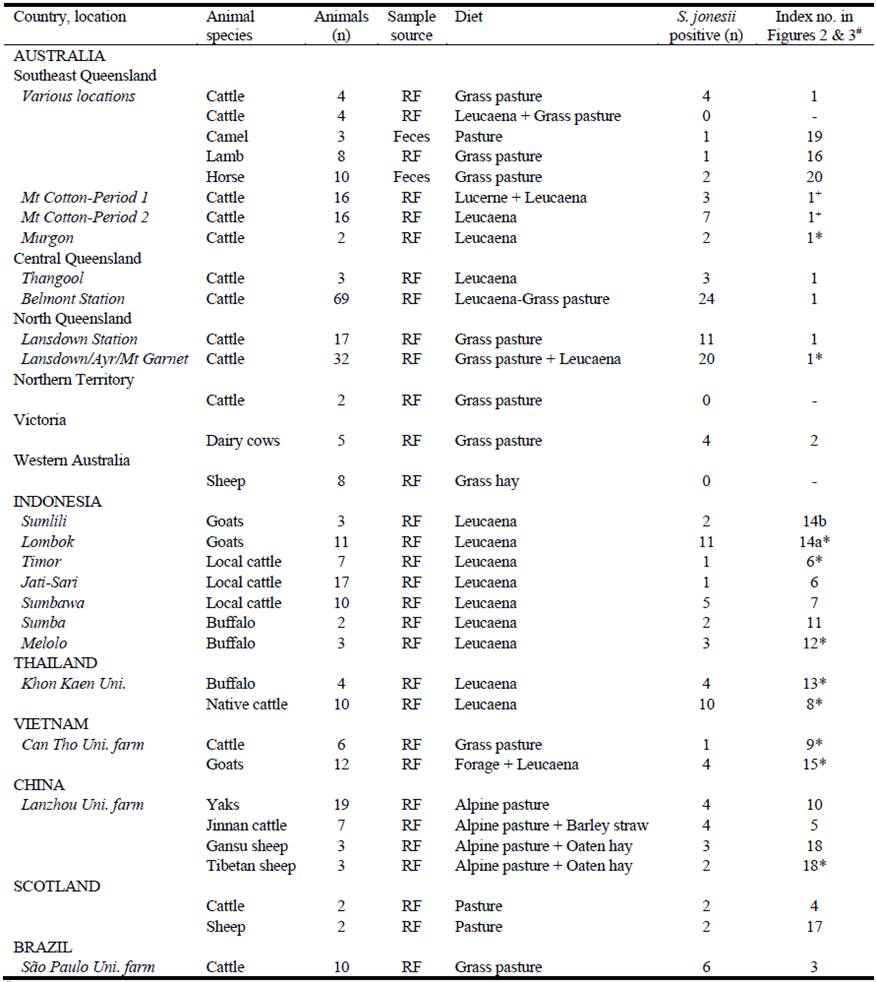
# Index number corresponds to sample number in parenthesis used in Figures 2 and 3 for SNP analysis.
S. jonesii positive rumen samples from Padmanabha et al. 2014* and Halliday et al. 2018+. RF= rumen fluid.
Detection of Synergistes jonesii by RT-qPCR
The RNA-based RT-qPCR increased the sensitivity for detection of S. jonesii by more than 100-fold compared with gDNA analysis in mixed cultures of rumen bacteria containing the target bacterium (Figure 1). The majority of rumen samples from 2 groups of cattle in Indonesia and 1 group in Australia, which tested negative for S. jonesii based on gDNA analysis (nested PCR), were mainly positive when RNA-based analysis was used (Table 3). This confirmed that the populations of S. jonesii were often at the limits of detection for nested PCR.
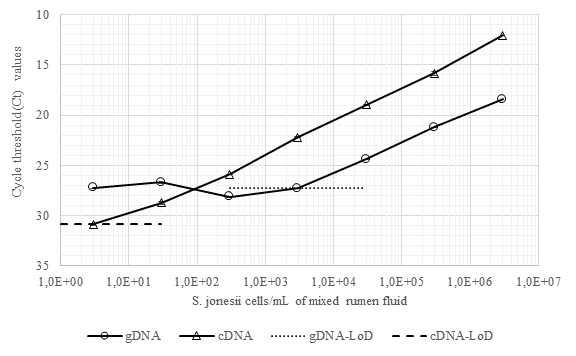
Figure 1 Detection and quantitation of S. jonesii cells spiked into mixed rumen culture by qPCR (using gDNA) and RT-qPCR (using cDNA). LoD = limit of detection.
Survey of SNP diversity in S. jonesii 16S rDNA
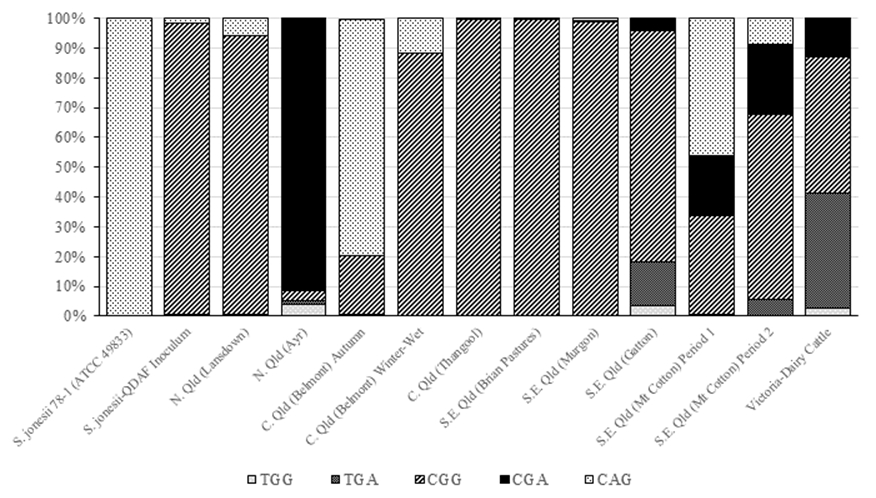
Synergistes jonesii type strain 78-1 contains the SNP variant CAG within the 16S rDNA. The Queensland Department of Agriculture and Fisheries (QDAF) inoculum, that is provided to cattle producers to protect their herds from toxicity, contains a CAG variant as a minor member while the dominant variant (98%) contains the CGG SNP pattern (Figure 2). The majority of livestock samples contained 3 or more of the SNP variants irrespective of their country of origin, while the remaining samples had 2 variants. Samples which did not contain the type strain (78-1) SNP included: Australian dairy cattle; camels and horses; some Chinese cattle and yaks; some Indonesian cattle and goats; and Vietnamese cattle.
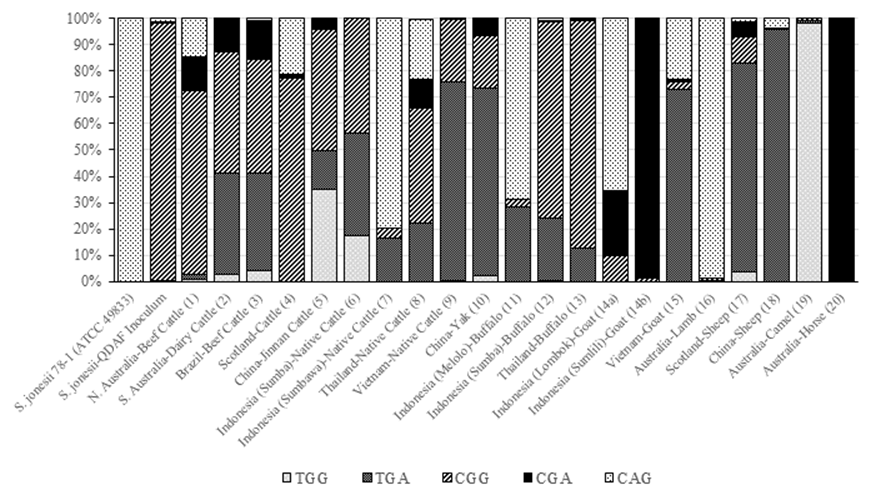
Figure 2 Distribution of S. jonesii 16S rDNA-specific SNPs in livestock species from different countries. The index numbers in parenthesis identify the samples from Table 2 used in the SNP analysis.
When Australian cattle were compared, the most common SNP pattern across animals was CGG, which is also the dominant SNP in the QDAF inoculum (Figure 3).
Discussion
The results of this study support the recent discovery that S. jonesii appears to be ubiquitous in ruminants rather than isolated geographically (Padmanabha et al. 2014). The nested PCR was able to detect S. jonesii usually at low numbers (<106 cells/mL) in most of the Australian cattle groups and overseas ruminants (cattle, buffalo, goats, sheep and yaks), whether feeding on leucaena or not, suggesting that the bacterium is indigenous to many of these animals and regions. Clearly, the RNA-based RT-qPCR method increased the sensitivity of detection of S. jonesii and would provide a more accurate measure of the prevalence of the bacterium in future surveys provided the rumen digesta-RNA can be immediately preserved upon collection using stabilizing chemicals/agents. Despite the increase in sensitivity, RNA-based PCR is not suitable for quantification of S. jonesii as it is based on an unknown number of target 16S rRNA gene copies per bacterium that can vary significantly depending on the stage and rate of growth of the bacterium. However a qualitative assessment of relative abundance could be achieved by performing a dilution series of the rumen fluid and noting the greatest dilution at which a positive test was observed. Increasing the abundance of S. jonesii in the rumen by providing peptides or specific amino acids which are required by the bacterium (Allison et al. 1992) has not been studied but should be examined in future as a way of increasing DHP degradation.
Although the indigenous nature of S. jonesii would imply inoculation is not required to transfer the microbe, the lack of complete degradation of DHP observed in many animals globally suggests S. jonesii alone is incapable of fully protecting ruminants on high leucaena diets. A recent review of leucaena toxicity in ruminants concluded that hepatic conjugation of DHP plays a major role in protecting animals from toxicity of DHP isomers that have not been completely degraded to non-toxic products in the rumen (Shelton et al. 2019). Although S. jonesii appears to be ubiquitous, work prior to the development of sensitive and specific molecular PCR assays reported that DHP degradation by rumen microbiota (presumably S. jonesii) did not occur in all countries tested (Jones 1984). Collectively these observations indicate that, while S. jonesii may be distributed widely in ruminants, their ability to degrade DHP could vary between animals and geographical regions.
The nested PCR amplicons from rumen samples identified as positive for S. jonesii were 454-sequenced to determine any variations of the 16S rDNA. Previously, longer (~800 bp) Sanger-sequencing analysis of the S. jonesii positive 16S rDNA sequences revealed 4 loci with point mutations at base positions (based on E. coli 16S rDNA numbering) 268 (C/T), 306 (A/G), 328 (G/A) and 870 (A/C) (Padmanabha et al. 2014). These are single nucleotide polymorphisms or SNPs, and, when present, occurred predominantly at loci 306 and 870 (Padmanabha et al. 2014). To facilitate a deeper analysis of the frequency of these SNPs in individual rumen samples, we designed PCR primers that would amplify a region that included only the first 3 of the 4 loci (~37‒380 bp), as a larger amplicon that included the locus at base 870 would not be amenable to 454-pyrosequence analysis. The SNP analysis demonstrated that the S. jonesii population in livestock is usually composed of several strains and never represented solely by the type strain (78-1) SNP (CAG).
The presence of SNPs within the 16S rDNA of the S. jonesii species implies there is genetic variation between closely related strains. This strain diversity may account for some of the differences observed in degradation of the 2 DHP isomers between animals. The total amount of undegraded DHP in the urine is most likely correlated with the amount of leucaena in the diet. It is possible that the DHP-degrading ability of different strains may vary, particularly if some S. jonesii bacteria are present in ruminants where leucaena and DHP are absent from the environment. Previous studies have demonstrated that the enzymes involved in the isomerization of 3,4-DHP to 2,3-DHP and cleavage of the pyridine ring are regulated by the concentration of pyridinediols (Rincón et al. 2000). The genetic regulation of these metabolic processes may have evolved differently depending on the rumen environment in which the bacteria reside. In a recent study, where cattle were dosed with the QDAF enrichment inoculum, Halliday et al. (2018) provided some evidence that there are differences in the DHP-degradation pathway between strains of these bacteria. They concluded that there were indigenous strains of S. jonesii in cattle, which converted 3,4-DHP to 2,3-DHP at a faster rate than 2,3-DHP was able to be degraded. Following inoculation, the extent of 2,3-DHP degradation appeared to increase and total DHP excretion decreased, indicating that the inoculum may have been more effective in degrading 2,3-DHP than the indigenous strains. Therefore despite already harboring indigenous S. jonesii, the provision of the QDAF inoculum may further enhance the ability of the rumen to degrade DHP. However it is also possible that other species of DHP-degrading bacteria, which contribute to these differences in metabolism of the two DHP isomers, could be present in the rumen (Allison et al. 1990; Domínguez-Bello et al. 1997).
In conclusion, S. jonesii was present as a minor population (<106 cells/mL) in the rumen of most animals tested and was usually comprised of several genetic variants of the species. Although the indigenous nature of S. jonesii could imply animals are protected from toxicity, the relative abundance of the bacterium, potential variation in DHP-degrading ability of genetic variants of the species, and level of intake of leucaena may all influence the capacity of the resident population in the rumen to protect the animal from toxicity.