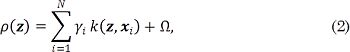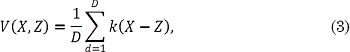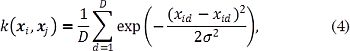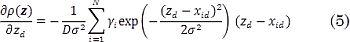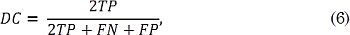Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
TecnoLógicas
Print version ISSN 0123-7799On-line version ISSN 2256-5337
TecnoL. vol.20 no.39 Medellín May/Aug. 2017
Artículo de investigación/Research article
Enhancement of nerve structure segmentation by a correntropy-based pre-image approach
Mejoramiento de la segmentación de estructuras nerviosas usando un enfoque de pre-imágenes basado en correntropía
J. Gil-González1, A. Álvarez-Meza2, J. Echeverry-Correa3, A. Orozco-Gutiérrez4, and M. Álvarez-López5
1 MSc in Electrical Engineering, Electrical Engineering Program,Universidad Tecnológica de Perei-ra, Pereira-Colombia, jugil@utp.edu.co
2 PhD in Engineering-Automatics, Electrical Engineering Department, Universidad Tecnológica de Pereira, Pereira-Colombia, andres.alvarez1@utp.edu.co
3 PPhD in Engineering-Electronic Systems, Electrical Engineering Department, Universidad Tecno-lógica de Pereira, Pereira-Colombia, jde@utp.edu.co
4 PhD in Bioengineering, Electrical Engineering Department, Universidad Tecnológica de Pereira, Pereira-Colombia, aaog@utp.edu.co
5 PhD in Computer Science, Department of Computer Science, University of Sheffield, Shef-field-United Kingdom, mauricio.alvarez@sheffield.ac.uk
Fecha de recepción: 03 de abril de 2017/ Fecha de aceptación: 03 de mayo de 2017
Como citar / How to cite
J. Gil-González, A. Álvarez-Meza, J. Echeverry-Correa, A. Orozco-Gutiérrez and M. Álvarez-López, “Enhancement of nerve structure segmentation by a correntropy-based pre-image approach”, TecnoLógicas, vol. 20, no. 39, 2017.
Abstract
Peripheral Nerve Blocking (PNB) is a commonly used technique for performing regional anesthesia and managing pain. PNB comprises the administration of anesthetics in the proximity of a nerve. In this sense, the success of PNB procedures depends on an accurate location of the target nerve. Recently, ultrasound images (UI) have been widely used to locate nerve structures for PNB, since they enable a non-invasive visualization of the target nerve and the anatomical structures around it. However, UI are affected by speckle noise, which makes it difficult to accurately locate a given nerve. Thus, it is necessary to perform a filtering step to attenuate the speckle noise without eliminating relevant anatomical details that are required for high-level tasks, such as segmentation of nerve structures. In this paper, we propose an UI improvement strategy with the use of a pre-image-based filter. In particular, we map the input images by a nonlinear function (kernel). Specifically, we employ a correntropy-based mapping as kernel functional to code higher-order statistics of the input data under both nonlinear and non-Gaussian conditions. We validate our approach against an UI dataset focused on nerve segmentation for PNB. Likewise, our Correntropy-based Pre-Image Filtering (CPIF) is applied as a pre-processing stage to segment nerve structures in a UI. The segmentation performance is measured in terms of the Dice coefficient. According to the results, we observe that CPIF finds a suitable approximation for UI by highlighting discriminative nerve patterns.
Keywords: Nerve structure segmentation, ultrasond images, pre-images approximation, Correntropy.
Resumen
El bloqueo de nervios periféricos (PNB) es una técnica ampliamente usada para llevar a cabo anestesia regional en el manejo del dolor. El PNB aplica una sustancia anestésica en el área que rodea el nervio que se quiere intervenir, y su éxito depende de la localización exacta del mismo. Recientemente, las imágenes de ultrasonido (UI) se han utilizado para la localización de nervios periféricos en PNB ya que permiten una visualización no invasiva y directa del nervio y de las estructuras anatómicas alrededor de él; sin embargo, este tipo de imágenes están afectadas por ruido speckle, dificultando su delimitación exacta. De esta manera, es pertinente una etapa de filtrado para atenuar el ruido sin remover información anatómica importante para la segmentación. En este artículo se propone una estrategia para el mejoramiento de UI usando filtrado basado en pre-imágenes. En particular, las imágenes se mapean a un espacio de alta dimensionalidad a través de una función kernel. Específicamente, se emplea un mapeo basado en Correntropía con el fin de codificar estadísticos de orden superior de las imágenes bajo condiciones no-lineales y no-Gaussianas. El enfoque propuesto se valida en la segmentación de nervios para PNB. El enfoque de filtrado basado en pre-imágenes con Correntropía (CPIF) es usado como pre-procesamiento en tareas de segmentación de nervios sobre UI. El rendimiento de la segmentación es medida en términos del coeficiente Dice. De acuerdo con los resultados, CPIF encuentra una aproximación adecuada para las UI al asegurar la identificación de patrones discriminativos de estructuras nerviosas.
Palabras clave: Análisis de componentes principales, Correntropía, Filtrado, Funciones Kernel, Segmentación.
1. Introduction
Ultrasound imaging is a widely used technique for visualization, analysis, and diagnosis of different areas in a human body [1]. In fact, UI offers several advantages compared to other types of medical imaging (e.g. magnetic resonance, x-rays, and computed tomography). The benefits include real-time imaging, no radiation, and small movable devices [2]. In particular, Ultrasound Images (UI) have been employed to locate nerve structures in Peripheral Nerve Blocking (PNB) procedures, and they facilitate pain management by regional anesthesia [3]. Since PNB comprises the administration of an anesthetic on the area surrounding a target nerve, an accurate localization of such region is crucial to ensure satisfactory results [4]. Therefore, this localization is a critical step for the success of PNB procedures as well [5]. In this regard, UI provides a direct visualization of relevant objects during PNB. They include the nerve, the anatomical structures around it (muscles, tissues, arteries and skin), the needle used during PNB, and the anesthetic drug [6]. However, this type of images are distorted by speckle noise that makes it difficult to accurately localize the region of interest [7]. Speckle noise is a form of multiplicative noise generated by the non-homogeneous structures analyzed in the tissue [8]. This noise degrades the image quality and affects the fine details and the edges of the anatomical structures [9]. Furthermore, another challenge during UI-based PNB comes from the dynamic properties of the nerve arrangement. The latter varies in shape and location, in line with the body position, the gravity, and the pressure of the ultrasound transductor [10]. Consequently, the success of PNB mainly depends on the operators expertise. They must define a proper setup for the ultrasound device, i.e., the gain, the spatial resolution, the axial resolution, the transductor position, and the contrast. As a result, the identification of nerve structures from UI is not a straightforward activity and it demands appropriate skills from anesthesiologists [4].
Recently, systems for computeraided nerve segmentation have been developed to assist the anesthesiologist throughout PNB [3]. Overall, such systems are based on automatic segmentation of nerve structures. This kind of segmentation presents the following open issues: i) the presence of speckle noise ii) the extraction of discriminative nerve patterns, and iii) the delimitation of nerve structures based on the extracted features. However, we highlight that speckle noise reduction is a critical step for nerve segmentation [11]. Speckle noise is a multiplicative noise generated by the heterogeneous structures of the tissue [8]. In fact, speckle noise significantly degrades the UI quality because it affects fine details and edges of the anatomical structures [9]. However, such noise may contain relevant information for the specialist, e.g. boundary sharpness to perform visual-based segmentation of anatomical structures [1].
In this sense, the problem of speckle noise reduction has been widely studied in the literature. Among the methods developed for speckle reduction we can find different approaches. Authors in [12] apply the Wiener filter to UI as data approximation using second-order statistics of a Fourierbased decomposition. However, this method is not adequate for speckle reduction since it is designed for reducing additive noise [13]. In turn, a homomorphic approximation was described in [14] to handle the multiplicative nature of speckle noise. Therefore, a logarithmic function is computed on the UI to map the multiplicative noise into an additive one. Then, a Wiener filtering approach is adopted. This method can effectively minimize speckle noise. Nevertheless, it fails to preserve useful anatomical details [13]. Moreover, authors in [15] introduce a multiresolution wavelet-based filtering that is implemented in the logarithmic mapping of the UI. Then, both the signal and noise components are modeled as Gaussian Processes to construct a Bayesian estimator for each scale and to support noise reduction and feature extraction stages. However, this approach assumes that the speckle noise can be mapped into additive Gaussian noise. Such assumption does not hold true in the context of UI, where the noise is likely to obey the Fischer-Tippet distribution [16].
Similarly, the median filter is a nonlinear filter that is commonly used to deal with multiplicative noise. This filter is very popular in the context of image processing due to its simplicity and noise reduction [13]. Yet, the median filter removes some high-frequency components and this produces a blurring effect at the edges [17]. On the other hand, authors in [18] present an extension of the Non-Local Means (NLM) filter proposed in [19]. This filter is based on the assumption that, for a given patch in the image, it is possible to find other patches with similar statistics. In this sense, the denoised patch will depend on both the pixels in the local patch and the pixels of the image with similar statistics. Nonetheless, the NLM filter was originally developed to deal with additive Gaussian noise. For this reason the authors employ a Bayesian framework to incorporate a model noise related to the UI features [18]. This filter, when adapted to UI, has shown interesting properties to remove speckle noise and preserve the edges [18].
Now, in the context of nerve identification from UI, a few studies have been proposed to deal with the particular features of nerve structures. Indeed, the filtering approach should enable UI enhancement to improve the segmentation by removing the components associated with speckle noise while highlighting the patterns related to nerve structures. For instance, authors in [20] introduce a segmentation approach based on Support Vector Machines (SVM) and Active Contour Segmentation (ACS). This approach involves the use of a linear filter (holding a homogeneous mask area) that is applied to minimize speckle noise. Similarly, in [21] the authors propose an approach based on active contours without considering a filtering step. Although promising results are obtained with sophisticated segmentation schemes, the above methodologies are not focused on speckle reduction.
In this paper, we propose an UI improvement strategy using a pre-image-based filter [22]. In particular, we map the input images by a nonlinear function (Kernel). We perform a principal component analysis within the feature space (Kernel PCA) to reveal important nerve properties and mitigate speckle noise perturbations. Then, based on the Kernel PCA analysis, we discard the directions with small variations. We hypothesize that these directions contain information related to speckle noise. Additionally, we employ a preimage method in order to retrieve the image in the input space. The main contribution of our work lies in the kernel functional, fixed as a correntropy-based mapping to code higher-order statistics from the input data under both nonlinear and non-Gaussian situations [23], e.g. multiplicative noise. The Correntropy-based kernel is similar to the NLM filter [18], [19] in the sense that both approaches analyze the similarities between different regions in the image. However, both methods have significant differences since NLM examines the gray level over square neigh-borhoods and the correntropy-based kernel performs this process pixel to pixel using an information theoretic learning-based similarity function [26]. So, our filtering approach is based on the preimage problem scheme that carries out subspace projection through correntropy-based kernel principal components analysis aiming to reveal significant nerve properties and to mitigate speckle noise perturbations. We validate our approach on a real-world UI dataset focused on nerve segmentation for PNB. In this regard, our Correntropy-based Pre-Image Filtering (CPIF) is applied as a pre-processing stage to further segment the nerve by using a Gaussian Processes strategy [4]. The obtained results show that CPIF finds a suitable UI approximation by ensuring the identification of discriminative nerve patterns. Indeed, our proposal outperforms state-of-the-art approaches that carry out UI filtering regarding a Dice coefficient assessment [24].
The remainder of this paper is organized as follows. In section 2 we describe the background of the introduced CPIF. In section 3 we present and discuss the nerve segmentation results including the CPIF-based UI enhancement. Finally, we outline the main conclusions and the future work in section 4.
2. Materials and methods
Pre-image problem: Let x be a point in the input space χ and let Ø(x) be its representation in the Reproducing Kernel Hilbert Space (RKHS) Hk associated with the kernel k(x,y)=Ø(x)TØ(y), where Ø(x) is a mapping function (usually non-linear) from the input space χ to the highdimensional (possibly infinite) space Hk. Let ψ ∈ Hk be a sample in the RKHS. The preimage approach finds its corresponding point x*∈ χ from 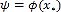 [25]. Since Hk is usually larger than χ and commonly we do not have acces to the map function Ø, an approximation of z can be computed by minimizing [25]:
[25]. Since Hk is usually larger than χ and commonly we do not have acces to the map function Ø, an approximation of z can be computed by minimizing [25]:
Thus, the PreImage approach has a significant range of applications in kernel methods, such as compression and denoising. In practice, we have a training set xi χ ℝ D, where: 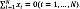 . Here, each sample xi corresponds to the vectorized version of the i–th UI. This way, to solve Eq. (1), a Kernel Principal Component Analysis (kPCA) is carried out to map the input data to Hk as a subspace projection based on the following eigenvalue problem: N λ a=Ka, where K ∈ ℝ N×N is a matrix holding elements Kij through the kernel function evaluation k(xi,xj), a ∈ ℝ N×N is an orthogonal eigenvector matrix, and λ ∈ ℝ N×N is a diagonal eigenvalue matrix where the eigenvalues are sorted in decreasing order. In this regard, kPCA constructs an orthogonal set of feature extractors in the RKHS as
. Here, each sample xi corresponds to the vectorized version of the i–th UI. This way, to solve Eq. (1), a Kernel Principal Component Analysis (kPCA) is carried out to map the input data to Hk as a subspace projection based on the following eigenvalue problem: N λ a=Ka, where K ∈ ℝ N×N is a matrix holding elements Kij through the kernel function evaluation k(xi,xj), a ∈ ℝ N×N is an orthogonal eigenvector matrix, and λ ∈ ℝ N×N is a diagonal eigenvalue matrix where the eigenvalues are sorted in decreasing order. In this regard, kPCA constructs an orthogonal set of feature extractors in the RKHS as 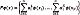 [22], where a ik represents the k-th eigenvector at the i-th position. Now, for a test image x*, the nonlinear components are extracted as
[22], where a ik represents the k-th eigenvector at the i-th position. Now, for a test image x*, the nonlinear components are extracted as 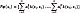 . In the case of denoising, we define an orthogonal set PM. This set is formed by the first M elements of the original set P, such as
. In the case of denoising, we define an orthogonal set PM. This set is formed by the first M elements of the original set P, such as 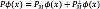 , where we assume that the discarded features extractors
, where we assume that the discarded features extractors 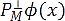 contain information associated with the speckle noise of the UI x. So, the filtered image in the RKHS is computed as PM Ø (x*) and, to calculate the pre-image z from PM Ø (x*), a denoised UI can be estimated by minimizing
contain information associated with the speckle noise of the UI x. So, the filtered image in the RKHS is computed as PM Ø (x*) and, to calculate the pre-image z from PM Ø (x*), a denoised UI can be estimated by minimizing 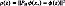 . Taking into account kernels of the form
. Taking into account kernels of the form 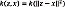 , the functional in Eq. (1) yields:
, the functional in Eq. (1) yields:
where 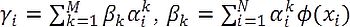 , and Ω is a term independent of z.
, and Ω is a term independent of z.
Overall, the Gaussian kernel is employed to perform preimage algorithms [22], [25]. However, this kernel is not a suitable choice in the context of speckle filtering, since such kind of noise does not follow a Gaussian distribution [16]. In this sense, as kernel functional, we employ a correntropy-based mapping to code higher-order statistics of the input data under both nonlinear and non-Gaussian situations. Correntropy is a measure that probabilistically estimates the similarities between two random variables [23]. This way, the correntropy functional generalizes the concept of correlation in nonlinear and non-Gaussian contexts, as follows:
where X,Z ∈ ℝ D, and k (X-Z) is a positive definite kernel. Fixing a Gaussian kernel in Eq. (3) allows describing the pixel-wise similarities between UIs, as follows:
where xid is the d-th pixel value of the UI xi, zd is the d-th pixel value of the filtered UI z, and σ² ∈ ℝ + is a kernel band-width. Consequently, a gradient descent approach is employed to solve Eq. (2) using a correntropy-based kernel, where the gradient w.r.t. z is given as:
In summary, the proposed pre-image filter computes a subspace projection in RKHS based on UI similarities from high-order dependencies using correntropy. Our filter also reduces the influence of speckle noise perturbations without affecting relevant patterns for nerve structure segmentation.
3. Results and discussion
3.1 Ultrasound imaging dataset
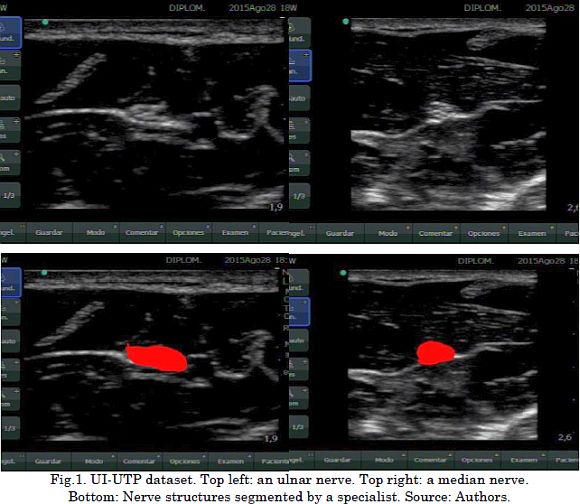
To validate the introduced Correntropy-based Pre-Image Filtering (CPIF), we use a dataset acquired by Universidad Tecnológica de Pereira and Hospital de Santa Mónica. This dataset is named UI-UTP and consists of UI recordings from patients who underwent regional anesthesia for PNB. This UI-UTP comprises 38 images: 16 from the ulnar nerve and 22 from the median nerve. Each UI was collected using a Sonosite Nano-Maxx device with a 640×480 pixel resolution. Each image in the dataset was labeled by one specialist in anesthesiology to indicate the location of the nerve structures. Fig.1 shows some UI-UTP dataset examples.
3.2 CPIF training and testing
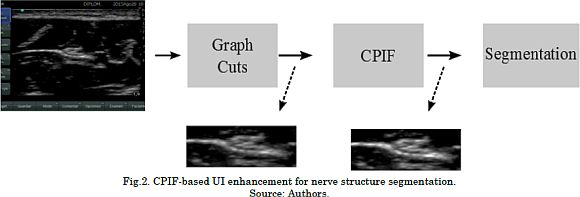
A leave-one-out validation scheme is employed to compute the system performance regarding the nerve segmentation after CPIF-based UI enhancement [20]. UI enhancement based on CPIF comprises the following stages: First, we use Graph Cuts segmentation [20] to define a region of interest (ROI) in which the nerve region is probably located. This ROI is necessary due to the fact that the number of pixels belonging to a nerve structure is much lower than the number of pixels in the image (Fig. 1). For concrete testing, ROI generation is carried out using the Graph Cuts algorithm [27], which allows to group the pixels into two clusters: background and object. Here, we use Graph Cuts because of their versatility to include topological constraints; for example, the nerves structures are not located on the edges of the UI. Now, since the ROIs hold different sizes, we perform a resizing step to ensure that all ROIs share the same pixel resolution. Then, CPIF is applied to the ROIs to reduce the speckle noise effect while enhancing the UI quality. The number of eigenvectors M and the correntropy free parameter σ² values are experimentally fixed from the leave-one-out strategy. Finally, these filtered ROIs feed the segmentation step that identifies the nerve structures. In this work, the segmentation step is carried out through a straightforward approach based on SLIC superpixels and Gaussian Processes, as described in [4]. Finally, the system performance is measured regarding the Dice coefficient (DC). The latter quantifies the overlap between the UI segment based on CPIF enhancement and the segment labeled by the specialist, as follows:
Where TP is the number of pixels that belong to the nerve that was classified as a nerve, FN is the number of pixels belonging to the nerve that were classified as background, and FP is the number of pixels that belong to the background that were classified as nerve. Namely, If there is a perfect segmentation, then DC=1. On the other hand, if there is not overlapping DC=0.
In addition, three state-of-the-art filtering methods are considered as benchmark: Non-Local Means [18], median filtering [13], and pre-image approach using a Gaussian kernel [22]. The free parameters for the Non-Local Means filter and the median filtering are experimentally fixed according to the applied validation scheme. Fig. 2 presents the main sketch of the introduced CPIF-based UI enhancements for nerve structure segmentation.
3.3 Obtained results
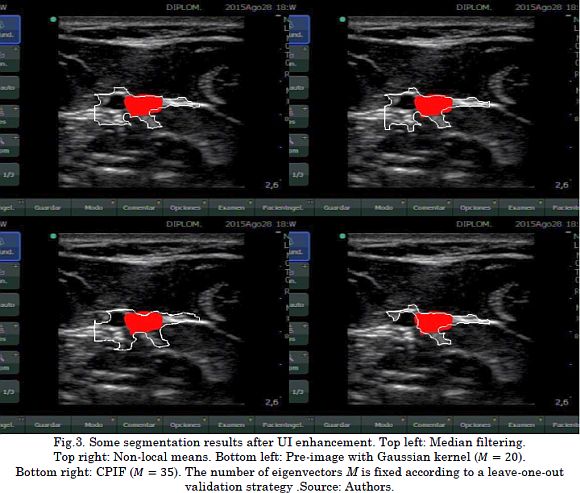
To visually compare the attained nerve identification after UI enhancement, Fig. 3 presents some segmentation results regarding each filtering approach They are median filtering, Non-Local Means, pre-image algorithm using a Gaussian kernel, and the introduced CPIF. In general, all these filtering methods allow highlighting significant patterns associated with nerve structures as long as the segmentation strategy identifies the nerves from the filtered UI. Nevertheless, median filtering, Non-Local Means, and pre-image approach based on Gaussian kernel obtain false positive segmentations related to other anatomical structures, e.g., blood vessels and the fascia. This is a significant issue because the specialist could supply the anesthetic in wrong places and cause possible complications, such as vessel damage or intoxication due to the introduction of the anesthetic drug into the bloodstream. Unlike these filtering methods, our CPIF approach considerably reduces the number of false positives, favoring a better representation of the nerves and the anatomical structures.
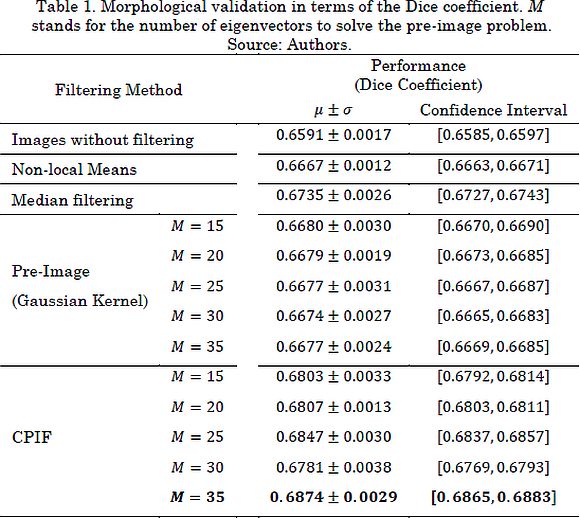
Now, Table 1 shows the results of the morphological validation in terms of the DC for the leave-one-out scheme. Based on the described results, our proposal outperforms state-of-the-art filtering methodologies. These results can be explained in the sense that CPIF offers a better representation of the composition of the image (i.e. the noise and the pattern associated with the anatomical structures). Therefore, CPIF allows a considerable reduction of speckle noise and highlights useful patterns employed for the identification of nerve structures. It is important to note that the number of retained eigenvectors plays an important role during UI enhancement. Since our pre-image-based approach depends on a correntropy similarity, the M value is bounded by the number of samples. Certainly, a tradeoff between data filtering and input information preservation must be considered when fixing such a value. In particular, in nerve structure segmentation tasks, the speckle noise may contain relevant information for the specialist (boundaries sharpness) [1].
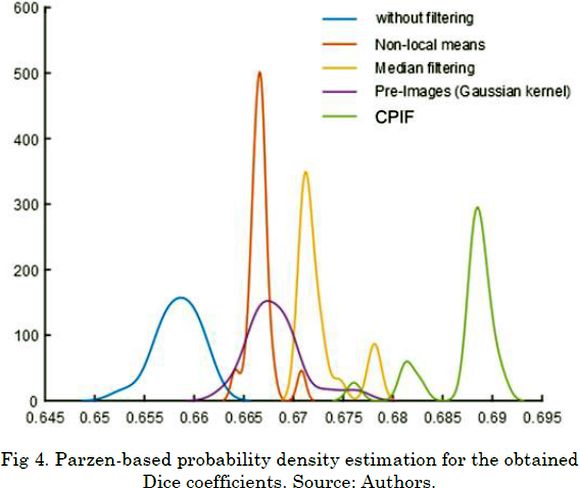
Now, to verify statistical differences among the studied preprocessing approaches, we estimate the probability density of the Dice coefficients using a Parzen-based nonparametric approximation. As seen in Fig. 4 , our CPIF approach outperforms state-of-the-art algorithms and obtains the highest Dice coefficient value with significant statistical differences concerning the probability densities. Indeed, our approach does not show significant overlaps with the probability densities of the others methods.
4. Conclusion
In this paper, we discuss a Correntropy-based Pre-Image Filtering (CPIF) to improve the segmentation of nerve structures depicted in UI. In this sense, a correntropy-based mapping is coupled within a pre-image approximation from RKHS to code higher-order statistics of UI. Thus, we introduce a gradient-based solution of the well-known pre-image filtering cost functional from a correntropy-based RKHS. As a result, nonlinear and/or non-Gaussian patterns related to nerve structures are enhanced to favor further segmentation tasks in PNB proce-dures. We tested our strategy on a real-world nerve segmentation dataset captured by the Automatics Research Group at Universidad Tecnológica de Pereira (Colombia). This dataset holds UI images of ulnar and median nerves. The experimental results showed that a CPIF that complements the raw UI by discarding noise components outperforms state-of-the-art alternatives for UI enhancement in terms of nerve segmentation accuracy. Hence, our CPIF-based filtering reduces the speckle noise effect without affecting relevant patterns used in the segmentation of nerve structures.
As future work, the authors plan to use more robust segmentation schemes to further improve the quality of the nerve segmentation, e.g., Support Vector Machine-based approaches [20]. Moreover, the introduced CPIF can be extended by adapting, not only the pre-image RKHS, but the approximation of the cost functional based on information theory measures [26].
5. Acknowledgments
J. Gil Gonzálezs work is funded by Colciencias under the program "Jóvenes Investigadores e Innovadores, Convocatoria 706 de 2015". Additionally, this work was funded by Colciencias under the project "Desarrollo de un sistema de identificación de estructuras nerviosas en imágenes de ultrasonido para la asistencia de bloqueo de nervios periféricos. Aplicación al tratamiento de dolor agudo traumático y prevención del dolor neuropático crónico" (código 1110-744-55958). Finally, we want to thank Dr. Diego Salazar from Hospital de Santa Mónica, who helped us to acquire the ultrasound images and to label the nerve structures.
References
[1] S. Sudha, G. R. Suresh, and R. Sukanesh, “Speckle noise reduction in ultrasound imag-es by wavelet thresholding based on weighted variance,” Int. J. Comput. theory Eng., vol. 1, no. 1, p. 7, 2009. [ Links ]
[2] R. Jirik and T. Taxt, “High-resolution ultrasonic imaging using two-dimensional homo-morphic filtering,” IEEE Trans. Ultrason. Ferroelectr. Freq. Control, vol. 53, no. 8, pp. 1440-1448, 2006. [ Links ]
[3] B. D. Sites and R. Brull, “Ultrasound guidance in peripheral regional anesthesia: philos-ophy, evidence-based medicine, and techniques,” Curr. Opin. Anesthesiol., vol. 19, no. 6, pp. 630-639, 2006. [ Links ]
[4] J. G. González, M. A. Álvarez, and Á. A. Orozco, “A probabilistic framework based on SLIC-Superpixel and Gaussian processes for segmenting nerves in ultrasound images,” in 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2016, pp. 4133-4136. [ Links ]
[5] N. M. Denny and W. Harrop-Griffiths, “Editorial I: Location, location, location! Ultra-sound imaging in regional anesthesia,” Br. J. Anaesth., vol. 94, no. 1, pp. 1-3, 2005. [ Links ]
[6] J. Shi, J. Schwaiger, and T. C. Lueth, “Nerve block using a navigation system and ul-trasound imaging for regional anesthesia,” in Engineering in Medicine and Biology So-ciety, EMBC, 2011 Annual International Conference of the IEEE, 2011, pp. 1153-1156. [ Links ]
[7] J. A. Noble and D. Boukerroui, “Ultrasound image segmentation: a survey,” Med. Im-aging, IEEE Trans., vol. 25, no. 8, pp. 987-1010, 2006. [ Links ]
[8] H. H. Choi, J. H. Lee, S. M. Kim, and S. Y. Park, “Speckle noise reduction in ultrasound images using a discrete Wavelet transform-based image fusion technique,” Biomed. Mater. Eng., vol. 26, no. s1, pp. S1587--S1597, 2015. [ Links ]
[9] H. Rabbani, M. Vafadust, P. Abolmaesumi, and S. Gazor, “Speckle noise reduction of medical ultrasound images in complex wavelet domain using mixture priors,” IEEE Trans. Biomed. Eng., vol. 55, no. 9, pp. 2152-2160, 2008. [ Links ]
[10] A. T. Gray, “Ultrasound-guided regional anesthesia,” Anesthesiology, vol. 104, no. 2, pp. 368-373, 2006. [ Links ]
[11] F. P. X. De Fontes, G. A. Barroso, P. Coupé, and P. Hellier, “Real time ultrasound im-age denoising,” J. real-time image Process., vol. 6, no. 1, pp. 15-22, 2011. [ Links ]
[12] W. K. Pratt, “Generalized Wiener filtering computation techniques,” IEEE Trans. Com-put., vol. 100, no. 7, pp. 636-641, 1972. [ Links ]
[13] M. C. Nicolae, L. Moraru, and L. Onose, “Comparative approach for speckle reduction in medical ultrasound images,” Rom. j. biophys, vol. 20, no. 1, pp. 13-21, 2010. [ Links ]
[14] S. Annadurai, Fundamentals of digital image processing. Pearson Education India, 2007. [ Links ]
[15] A. Achim, A. Bezerianos, and P. Tsakalides, “Novel Bayesian multiscale method for speckle removal in medical ultrasound images,” IEEE Trans. Med. Imaging, vol. 20, no. 8, pp. 772-783, 2001. [ Links ]
[16] O. V Michailovich and A. Tannenbaum, “Despeckling of medical ultrasound images,” IEEE Trans. Ultrason. Ferroelectr. Freq. Control, vol. 53, no. 1, pp. 64-78, 2006. [ Links ]
[17] Y. Chen, R. Yin, P. Flynn, and S. Broschat, “Aggressive region growing for speckle reduction in ultrasound images,” Pattern Recognit. Lett., vol. 24, no. 4, pp. 677-691, 2003. [ Links ]
[18] P. Coupé, P. Hellier, C. Kervrann, and C. Barillot, “Nonlocal means-based speckle filter-ing for ultrasound images,” IEEE Trans. Image Process., vol. 18, no. 10, pp. 2221-2229, 2009. [ Links ]
[19] A. Buades, B. Coll, and J.M. Morel, “A non-local algorithm for image denoising,” in Computer Vision and Pattern Recognition, 2005. CVPR 2005. IEEE Computer Society Conference on, 2005, vol. 2, pp. 60-65. [ Links ]
[20] O. Hadjerci, A. Hafiane, N. Morette, C. Novales, P. Vieyres, and A. Delbos, “Assistive system based on nerve detection and needle navigation in ultrasound images for re-gional anesthesia,” Expert Syst. Appl., vol. 61, pp. 64-77, 2016. [ Links ]
[21] A. Hafiane, P. Vieyres, and A. Delbos, “Phase-based probabilistic active contour for nerve detection in ultrasound images for regional anesthesia,” Comput. Biol. Med., vol. 52, pp. 88-95, 2014. [ Links ]
[22] S. Mika, B. Schölkopf, A. J. Smola, K.-R. Müller, M. Scholz, and G. Rätsch, “Kernel PCA and De-noising in feature spaces.,” in NIPS, 1998, vol. 11, pp. 536-542. [ Links ]
[23] S. Zhao, B. Chen, and J. C. Principe, “Kernel adaptive filtering with maximum corren-tropy criterion,” in Neural Networks (IJCNN), The 2011 International Joint Conference on, 2011, pp. 2012-2017. [ Links ]
[24] M.-J. Lesot, M. Rifqi, and H. Benhadda, “Similarity measures for binary and numerical data: a survey,” Int. J. Knowl. Eng. Soft Data Paradig., vol. 1, no. 1, pp. 63-84, 2008. [ Links ]
[25] G. H. Bakir, J. Weston, and B. Schölkopf, (2004). “Learning to find pre-images. [ Links ]”
[26] J. C. Principe, D. Xu, and J. Fisher, (2000). Information theoretic learning. Unsupervised adaptive filtering, 1, 265-319. [ Links ]
[27]Y. Boykov and G. Funka-Lea, (2006). Graph cuts and efficient ND image segmentation. International journal of computer vision, 70(2), 109-131. [ Links ]