Autoimmune diseases are pathophysiologically complex and multifactorial medical conditions that, even today have a big pending in relation to their clinical (diagnosis, prognosis, treatment) and basic (etiopathogenesis) understanding. Even though the practice of modern rheumatology has been enriched by the progressive arrival of more advanced methods of cellular and molecular studies, there are still shortcomings in the knowledge of the etiopathogenic processes of the diseases.
It is also true that the more is known about etiopathogenic phenomena and mechanisms of autoimmune diseases, the diagnostic and therapeutic arsenal is not only widened, but the need to continue learning stands out, since with the advent of biological drugs, new nosological conditions, for example, immune-mediated adverse events related to biological drugs such as checkpoint inhibitors have arisen.
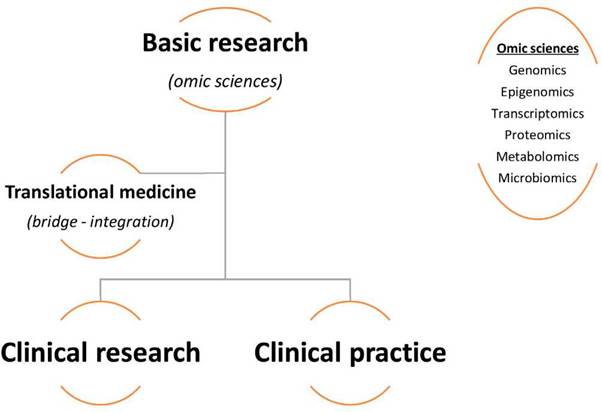
For all these reasons, an imperative need for information from the basic sciences arises, that clarifies many still unknown aspects of autoimmune diseases and directly brings basic knowledge closer to clinical practice.1 This is how translational medicine and its multiomic approaches arise, which in their fundamental structure (Fig. 1) get to strengthen knowledge in rheumatology and its clinical practice, given that its objective is to "transfer" directly and preferentially what is found in basic biological studies to the field of clinical medicine, being applicable not only to the treatment and diagnosis, but also to the prognosis and precision medicine, despite the research challenges to which this science is compelled (Fig. 2). Then, rheumatology has a great opportunity to benefit from translational medicine and its wide omic terrain (genomic, epigenomic, transcriptomic, proteomic, metabolomic, microbiomic), given the possible answers they can offer to the knowledge gaps of the specialty. (2
Source: own elaboration.
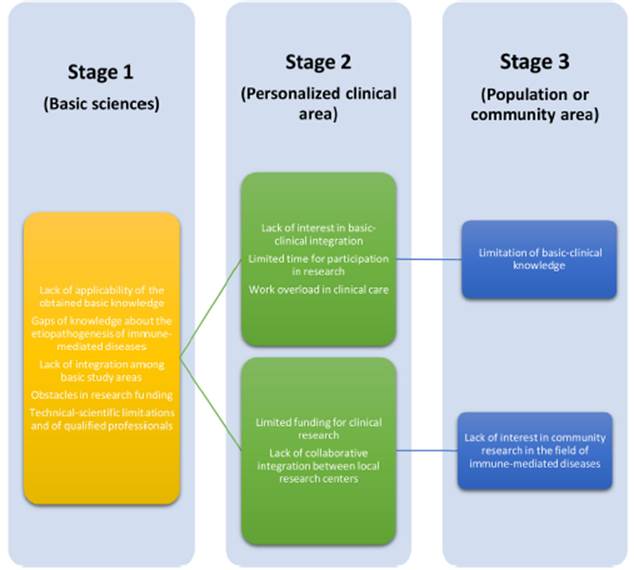
Fig. 2 Operational challenges in translational research in the field of rheumatology.
The large databases of global human genomic studies have seen how relevant populations such as the Latin American or African, and types of diseases such as autoimmune, lack of a significant representation in the studies, therefore, an effort must be made to solve this shortcoming.3,4
It is important to generate local own knowledge in relation to genetics and omic sciences, since Colombia is a country with a diverse ethnic population composition and mainlyconstituted by 3 groups: Amerindians, European immigrants and Afrodescendants.5 For this reason, in this issue of the journal we highlight two Colombian studies that provide knowledge in the field of omic sciences (genetics/genomics) and autoimmune diseases.
Thanks to the contribution of Arévalo-Caro et al., it has been determined that in Colombia, as in other latitudes, the genetic association between the alleles of the HLA-DRB1 gene known as "shared epitope" and rheumatoid arthritis (RA) in mestizo subjects of Bogota presented an equivalence of 100%, highlighting the presence of HLA-DB1 alleles 14:02, 04:04, 08:02, 04:05 and 10:01 in the patients with RA and pointing out the higher frequency of the shared epitope in subjects with RA and anti-cyclic citrullinated peptide antibodies. On the other hand, Garavito et al. stated that the polymorphims of the vitamin D receptor gene, such as the VDR rs2228570 and the ACCA haplotype of the Taql, Apal, Bsml and Fokl polymorphisms, were associated with an increased risk of systemic lupus erythematosus in a sample of adolescent subjects from the Colombian Caribbean.6,7
Like the academicians, leaders and researchers that usually are immersed, the specialists in the different clinical areas must try to overcome the barriers that condition the conduct of research at the level of translational medicine. Fortunately, we already visualize groups that fully comply with this principle and we invite everyone to join this cause to overcome the lag that we have at the local and regional level compared with the countries of the First World.











 text in
text in