Introduction
Antibiotic resistance is expanding rapidly throughout the world and has become a problem in both human and veterinary medicine (FDA, 2012; Reist et al., 2013). The emergence of this resistance in bacteria found in animals and their products has motivated considerable interest because of the potential for resistance transfer to the human population (McDermott et al., 2002).
One of the greatest concerns is the rapid increase of resistant bacteria that produce extended-spectrum beta-lactamases (ESBLs), which reduce the efficacy of a wide range of beta-lactam antibiotics, such as third- generation cephalosporins and monobactams (Philippon et al., 1989; Bush, 2010). This resistance is based on genes of chromosomic or plasmidic origin, which encode enzymes that inactivate these compounds by hydrolysis of their beta-lactam ring (Paterson and Bonomo, 2005; Reist et al., 2013). These genes can be easily transferred between bacteria through mobile genetic elements, which often carry additional genes for resistance to other groups of antibiotics, compromising the efficacy of treatments for infections caused by resistant bacteria (Smet et al., 2010).
Until the nineties, the most prevalent ESBLworldwide were Temoniera (TEM) and sulfhydrylvariable (SHV) type associated mainly to hospital outbreaks caused by Klebsiella pneumoniae. However, since 2000, the enzyme cefotaxime-munich (CTX-M) became one of the most frequent and ESBL-producing Eschericha coli emerged as an important pathogen in the community (Blanco et al., 2016).
The CTX-M isolates have been found in various species of Enterobacteriaceae from different geographic areas: mainly during nosocomial outbreaks that occurred in Japan, Europe, South America, Africa, China, Korea and the United States (Villegas et al., 2004). In 2003, a study conducted in three Colombian hospitals located in Bogotá, Medellín, and Cali detected isolates from K. oxytoca, K. pneumoniae, and E. coli that harbored CTX-M enzymes. That study confirmed that CTX-M enzymes also occur in Colombia (Villegas et al., 2004). Further studies in Colombia indicated that this antibiotic resistance patterns from several hospital isolates have spread, especially in Gram-negative bacteria (Sánchez et al., 2008; Villegas et al., 2011; Leal et al., 2013; Amado et al., 2014; González and Cortés, 2014; Blanco et al., 2016).
The entry of ESBL-producing Enterobacteriaceae in the food chain and environment could be considered a possible interface for the exchange of resistance genes between humans and animals (WHO, 2001; Walsh and Fanning, 2008; EFSA, 2011). Raw milk, for example, can be contaminated intramammary with Enterobacteriaceae during mastitis processes, directly through animal feces or indirectly during milking through milkers or milking equipment (Dahmen et al., 2013), and also by cow contact with feces in the farm environment such as pens (Ferens and Hovde, 2011). However, all pathogenic microorganisms are destroyed after pasteurization, including multi-resistant bacteria, making milk and other dairy products generally safe for human consumption (Leedom, 2009; Lejeune and Rajala-Schultz, 2009). Raw milk is still used by a large number of rural families and a growing segment of the population who believe raw milk is generally safe and has beneficial health effects that are destroyed by pasteurization (Zeinhoma et al., 2014). According to data from the Ministerio de Agricultura y Desarrollo Rural de Colombia, 59% of the milk produced in the country is intended for commercialization through intermediaries, processed, and used for self-consumption, among others, and the remaining 41% is processed by the dairy industry (Decreto 1880, 2011, Ministerio de la Protección Social). Thus, the transmission of ESBL-producing Enterobacteriaceae to humans can occur through direct contact and/or ingestion of foods contaminated with ESBL-producing strains, among other routes (Wooldridge, 2008; Verraes et al., 2013).
Resistance to wide-spectrum antibiotics such as cephalosporins is increasing among food-producing animals, and food of animal origin can be considered a possible reservoir of ESBLs (Mesa et al., 2006; Xian-Zhi et al., 2007; Carattoli, 2008; Leverstein-van Hall et al., 2011). The excessive or inappropriate use of antibiotics in farm animals can generate adaptation of bacteria to antimicrobials (FAO, 2011); thus, it has become a matter of great importance for health authorities (WHO, 2001; EFSA, 2011).
The presence of ESBL-producing Enterobacteriaceae in dairy animals in Colombia is unknown. Although there are policies restricting the sale of antimicrobials, com pliance is minimal, which facilitates the appearance of resistance (Machado-Alba and González-Santos, 2009; Vacca et al., 2011). Thus, it is necessary to investigate the frequency of multi- resistant bacteria in foods of animal origin and to establish the possible role of dairy farms in their dissemination (Smet et al., 2010; Timofte et al., 2014).
This study aims to characterize the presence and frequency of ESBL-producing Enterobacteriaceae from bulk-tank milk samples of dairy farms in Entrerríos (Antioquia, Colombia), and explore potential risk factors associated with these bacteria in bulk-tank milk.
Materials and methods
Location
This study was conducted in Entrerríos, municipality in Northern Antioquia (Colombia), which is the main dairy region of this province. Average temperature in Entrerríos is 16 °C. The altitude is 2,300 m.o.s.l. and is located approximately 60 Km North of Medellín city, the capital of Antioquia province. Dairy farming is the most important economic activity in the municipality, which is located close to the metropolitan area of the Valle de Aburrá (Gobernación de Antioquia, 2013). The average daily milk production is 238,854 L (Gobernación de Antioquia, 2007). Most milk is sold to a couple of dairy processors and the rest is destined for domestic consumption and feeding of farm animals (Gobernación de Antioquia, 2013).
Selection of farms
From a total of 952 dairy farms in Entrerríos (based on year 2013 foot-and-mouth disease vaccination records from the Secretaría de Asistencia Tecnica y Desarrollo de Entrerrios), 120 farms were randomly selected. This number was based on an expected frequency of 0.10 at the farm level (Odenthal et al., 2013), a 90% confidence level and a maximum acceptable error rate of 0.10. The number of dairy farms to be sampled from each district was determined by proportional allocation. Only farms having their own bulk-tank were selected. That is, farms that store milk in collective or community tanks were not considered. An additional list of possible farms was randomly selected to replace farms where milk had already been collected by the truck on the sampling day.
Sample collection
Bulk-tank milk samples were collected in September and October, 2013. Each sample (30 mL) was obtained directly from the tank in a sterile container, immediately cooled and transported to the laboratory (Toro, 2012), where they were kept refrigerated and processed within 24 h of sampling.
Isolation and identification of ESBL-producing Enterobacteriaceae
For selective isolation (screening) of ESBL- producing bacteria, 100 μL of milk was spread onto chromogenic agar chromID™ ESBL (bioMérieux, Marcy-l’Étoile, France), which was incubated at 37 ºC for 20 to 24 h. Samples that displayed colony growth with pink-burgundy, green, blue or brown colorations were considered presumptive ESBL- producing Enterobacteriaceae. In each sample, one colony of each coloration was taken and spread onto McConkey agar (bioMérieux, Rio de Janeiro, Brazil) for subsequent identification. Each isolate was weekly replicated in McConkey agar for preservation. The form of the colonies was compared between samples with growth of various colonies of the same coloration, and two colonies of different morphology were taken. The commercial BD BBL™ Crystal™ Enteric/ Nonfermenter ID Kit (E/NF, Sparks, MD, USA) was used according to the manufacturer instructions to identify genus and species of Enterobacteriaceae. Only glucose fermenting colonies were selected, whereas the non-fermenting colonies were discarded.
Confirmation of ESBL production
The ESBL production was confirmed based on the double disk synergy test. For this, Mueller- Hinton agar (bioMérieux, Rio de Janeiro, Brazil) was inoculated with a bacterial suspension of 0.5 turbidity pattern in the McFarland standard, and discs of cefotaxime (CTX, 30 μg), ceftazidime (CAZ, 30 μg), ceftriaxone (CRO, 30 μg), cefepime (FEP, 30 μg), and aztreonam (ATM, 30 μg) were placed 20 mm center to center from a central disk of amoxicillin/ clavulanic acid (AMC). The agar inoculated with a bacterial suspension (Mc Farland turbidity pattern of 0.5) and discs were incubated at 37 °C for 18-24 h. An increase in the zone towards the AMC was considered positive for ESBL production according to criteria by the Clinical and Laboratory Standards Institute (CLSI 2014, Document M24). Reference strains of E. coli American Type Culture Collection (ATCC) 25922 and K. pneumonieae ATCC 700603 were used as negative and positive controls, respectively, in the double disk synergy test.
Antimicrobial susceptibility test
The antimicrobial susceptibility test was performed by the disc diffusion method in agar, according to recommendations by CLSI (2015). For this purpose, bacterial colonies were suspended in brain heart broth (bioMérieux, Marcy-l’Étoile, France) and adjusted to a McFarland turbidity pattern of 0.5 to be inoculated in a plate of Mueller-Hinton agar. Discs of the following antibiotics were evaluated: ampicillin (10 μg), norfloxacin (10 μg), imipenem (IPM, 10 μg), amoxicillin (10 μg), cefoxitin (FOX, 30 μg), cefuroxime (30 μg), ciprofloxacin (CIP, 30 μg), chloramphenicol (30 μg), doxycycline (30 μg), gentamicin (GEN, 10 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), erythromycin (15 μg), and cephalexin (30 μg; Oxoid, Basingstoke, Hampshire, England). After incubation for 18-24 h, inhibition halos (margin of the zones where no visible growth was observed) were measured in millimeters and interpreted as sensitive, intermediate or resistant according to CLSI (2015) standards.
Determination of minimum inhibitory concentration (MIC)
Antimicrobial MIC for isolates of ESBL-producing Enterobacteriaceae was determined by the broth microdilution method using the Vitek 2 07.01 system (bioMérieux, Marcy-l'Étoile, Francia). A total of 14 antibiotics were evaluated and interpreted according to CLSI (2008) standards: ampicillin/sulbactam (SAM), FOX, CAZ, CRO, FEP, doripenem (DOR), ertapenem (ETP), IPM, meropenem (MEM), amikacin (AMK), GEN, CIP, tigecycline (TGC), and colistin (COL).
Characterization of ESBL by polymerase chainreaction (PCR)
Positive ESBL isolates were additionally analyzed by PCR for the presence of bla genes of ESBL subtypes TEM, SHV, and CTX-M (groups 1, 2, 8, 9, or 25) using primers and conditions previously described (Pitout et al., 1998; Batchelor et al., 2005; Woodford et al., 2006). Bacterial DNA was isolated with the innuPREP bacteria DNA kit (Analitykjena, Berlin, Germany) according to the manufacturer’s instructions. Two strains, K. pneumoniae ATCC 700603 (which harbors a blaSHV gene) and an isolate of K. pneumoniae (which harbors both blaCTX-M and blaTEM genes) from the strain collection of the Department of Milk Sciences of the University of Giessen were used as positive ESBL standard strains. An ESBL non-producing strain E. coli ATCC 25922 was used as negative control. PCR products were determined by electrophoresis in a 2% agarose gel (Biozym, Hessisch Oldendorf-, Germany). The Gene Ruler 100 bp DNA molecular marker (MBI Fermentas, St. Leon-Roth, Germany) was used.
Sequencing of bla genes
Genes coding for ESBL bla TEM, bla SHV, and bla CTX-M of ESBL-positive isolates were amplified with primers and PCR conditions as described
previously (Pitout et al., 1998; Batchelor et al., 2005). The resulting amplicons were purified using the QIAquick PCR Purification kit (Qiagen, Hilden, Germany). Sequencing was performed in Seqlab (Goettingen, Germany). The results were evaluated using the BLAST algorithm available at http://blast. ncbi.nlm.nih.gov/Blast.cgi.
Collection of farm information and management practices
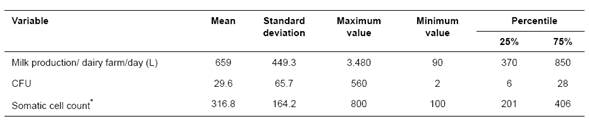
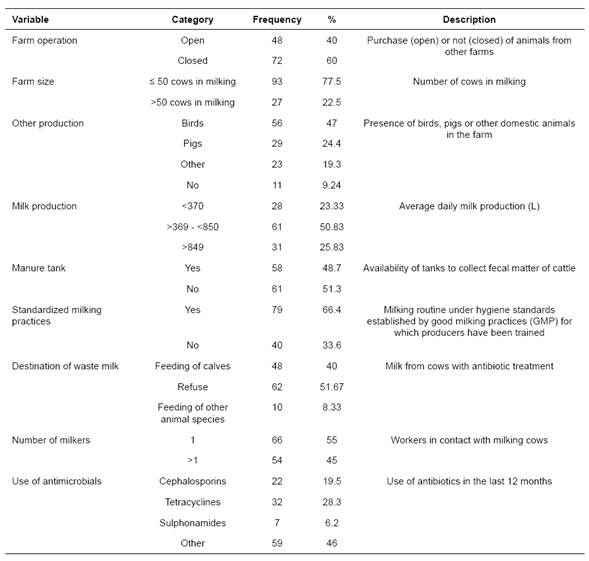
Information on the characteristics and management practices that could be associated with the presence of ESBL-producing Enterobacteriaceae was obtained using a questionnaire (supplemental material) at the time of bulk-tank sampling. Information on variables related to general conditions of the farm, production, sanitation, and hygiene, and the use of antibiotics was collected (Tables 1 and 2). Information about CFU values was obtained from the purchase invoice generated by the milk processor. Farms were classified as small (≤ 50 cows in milking) and large (>50 cows in milking).
Case definition
Bulk-tank milk was considered the unit of analysis. A tank was considered positive for the presence of ESBL-producing Enterobacteriaceae when at least one (1) isolate of Enterobacteriaceae was confirmed as an ESBL producer.
Statistical analysis
Data were stored in Excel 2010 (Microsoft, Redmond, USA) spreadsheets and later exported to Stata 12.0 (StataCorp, Texas, USA) for statistical analysis. Data were initially analyzed by descriptive statistics through measures of central tendency and frequency distribution. Bivariate logistic regression analysis was performed to evaluate association between variable, presence of ESBL in the tank, and some predictors of interest. Associations with p<0.25 were considered significant and were selected for the multivariate model. In the final model, only those variables with p<0.05 remained.
Results
General characteristics of the farms
The main characteristics are summarized in Tables 1 and 2. In total, 60% (72/120) of the sampled farms operated as closed farms; 77.5% (93/120) had at least 50 cows in milking. In addition, 91% (108/120) had animals of non-bovine species, with the presence of domestic birds and pigs in 47% (56/119) and 24.4% (29/119) of the farms, respectively. Moreover, 51.3% (61/119) of the farms had no manure tank, and 66.4% (79/119) reported performing the milking routine following technical recommendations that were focused on good livestock practices. Feeding calves waste milk was practiced in 40% (48/120) of the farms. A 55% (66/120) had one milker and 45% (54/120) of the farms had more than one milker.
A total of 70% (84/120) farms reported antibiotic usage formulated by a veterinarian only 50% of the times or less. Average milk production was 659,000 ± 449 L/day. Additionally, in 75% (90/120) of the farms, counts of colony forming units (CFUs) and somatic cells were below 28.000 CFU/mL and 406,000 cells/ mL, respectively (Table 2).
Characteristics of dairy farms with confirmed ESBL-producing isolates
In total, 75% (3/4) of the farms with isolates positive for ESBL production operated as closed farms, 4 farms had >50 (average 93) cows in milking, 100% of the farms also had horses and pets (dogs and cats), 50% (2/4) of the farms reported the presence of birds, while pigs were reported in only one farm. Additionally, 75% (3/4) of the farms had a manure tank, and the number of milkers was different among positive farms (1, 3, or 5 milkers). Calf feeding with waste milk was reported in 100% of the positive farms, and milking was performed under technical recommendations in 75% of those farms. The use of antimicrobials to treat animals following veterinary recommendations was reported as usually in one farm, sometimes in two farms, and rarely in more than two farms. The most frequently used antimicrobials in the past year in these four farms included penicillins (75%, 3/4), macrolides (50%, 2/4), and tetracyclines and quinolones (25%, 1/4). Those four farms produced 1,865 L/day. The CFU and somatic cell counts were 39,000 CFU/mL and 401,000 cell/mL on average.
Presence of ESBL-producing Enterobacteriaceae
Bacteria suspicious of producing ESBL were isolated in 6.6% (8/120) from milk samples of equal number of bulk-tanks. One isolate was obtained in each of the 8 tanks. Of these 8 isolates, 2 non- fermenting glucose isolates were discarded. Of the 6 remaining isolates, ESBL production was confirmed in 4; thus, the apparent frequency of ESBL-producing Enterobacteriaceae in bulk-tank milk was 3.3% (4/120; CI95%: 3-3.5%). All of the isolates showed synergy with at least two of the cephalosporins used and clavulanic acid. The isolates of ESBL- producing Enterobacteriaceae were identified as Enterobacter cloacae, Serratia fonticola, E. coli, and Enterobacter cancerogenus using the Crystal™ Enteric/Nonfermenter ID kit (E/NF; Becton Dickinson (BD) Franklin Lakes, New Jersey, USA).
Table 1 Selected predictors for exploring risk factors associated with the presence of ESBL-producing Enterobacteriaceae in bulk-tank milk (n = 120) at Entrerríos, Antioquia (Colombia).
Antimicrobial susceptibility
All four ESBL-producing isolates were resistant to ampicillin, amoxicillin, cephalexin, cefuroxime, erythromycin, and trimethoprim/sulfamethoxazole. Conversely, all four isolates were sensitive to FOX, IPM, CIP, and norfloxacin (Table 3). The isolate of E. coli was resistant to eight antibiotics (ampicillin, amoxicillin, cephalexin, cefuroxime, erythromycin, chloramphenicol, doxycycline, and trimethoprim/ sulfamethoxazole) and had the greatest resistance compared to the rest. The isolates of S. fonticola had the least resistance to the antibiotics (6/13) compared to the rest of the isolates and was the only isolate, in addition to E. cancerogenus, that displayed sensitivity to gentamicin. The S. fonticola and E. cloacae were the only isolates sensitive to chloramphenicol and doxycycline.
Minimum inhibitory concentration (MIC)
The MIC of 13 antibiotics obtained through the broth microdilution method in the 07.01 Vitek 2 (bioMérieux) for the four isolates is shown in Table 4. The only isolate that displayed sensitivity to SAM was E. coli, which had 8 mg/mL MIC. For the other three isolates, resistance or intermediate resistance level was observed. All isolates showed sensitivity to DOR, ETP, IPM, MEM, AMK, CIP, and TGC (Table 4). For CRO, all isolates yielded a MIC value that was interpreted as resistance. By contrast, for FEP and CAZ, only E. coli was resistant, with ≤ 1 mg/mL MIC (by automatic modification of the values obtained by the automated expert system Vitek® 2 (bioMérieux, Durham, NC, USA) and ≥ 64 mg/mL MIC, respectively.
The S. fonticola and E. cloacae isolates showed resistance to FOX, with 16 and ≤ 4 mg/mL MIC, respectively. The latter MIC was reported as resistant by automatic modification of values performed by the Vitek® system. The only isolate that showed resistance to GEN was E. cloacae with ≥ 16 mg/mL MIC. For the rest of the antibiotic groups, the MIC varied according to the isolated bacteria.
Table 3 Antimicrobial susceptibility patterns obtained by the disk diffusion method for four isolates of ESBL-producing Enterobacteriaceae in bulk-tank milk (n = 120) at Entrerríos, Antioquia (Colombia).
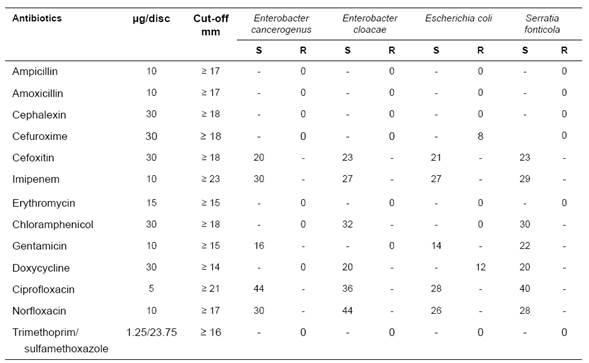
S: sensitive; R: resistant.
Table 4 Minimum inhibitory concentration (MIC) values obtained by the microdilution method in 07.01 Vitek 2 broth (bioMérieux) for 4 isolates of ESBL-producing Enterobacteriaceae in bulk-tank milk (n = 120) at Entrerríos, Antioquia (Colombia).
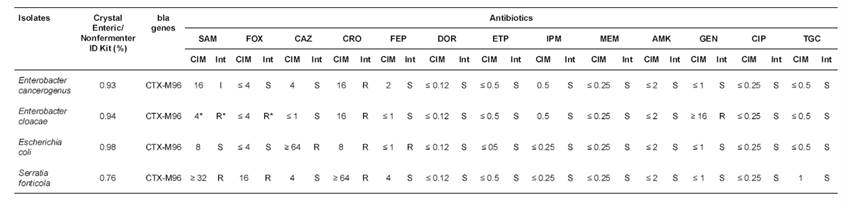
Ampicillin/sulbactam (SAM), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), doripenem (DOR), ertapenem (ETP), imipenem (IPM), meropenem (MEM), amikacin (AMK), gentamicin
(GEN), ciprofloxacin (CIP), tigecycline (TGC), colistin (COL).
MIC: minimum inhibitory concentration, Int: interpretation, S: sensitive, I: intermediate, R: resistant.
*values modified by the advanced expert system
Characterization of ESBL by PCR and sequencing of bla genes
The four isolates hosted CTX-M genes, based on PCR analysis. All of the isolates were positive for CTX-M -1 group, and when these were sequenced, all sequences showed strong homology with CTX-M 96 variant, which is alternatively called CTX-M-12a.
Risk factors
A univariate logistic regression analysis yielded an association between the presence of ESBL- producing Enterobacteriaceae and the size of the farm, and between the presence of ESBL-producing Enterobacteriaceae and milk production (Table 5). At the end, only farm size variable remained significant in the multivariable model constructed by backward elimination, p<0.05. The odds of having ESBL- producing Enterobacteriaceae in bulk-tank milk were 11.5-fold higher in farms with more than 50 cows in milking compared to farms equal or less than 50 cows in milking (p<0.038).
Discussion
The main limitations of this study were the absence of previous studies on ESBL-producing Enterobacteriaceae in bulk-tank milk from comparable dairy regions of Colombia, which would have estimated frequency according to national productive conditions, and a larger budget, which would allow larger sample size and different time periods. Taking into account the low number of positive isolates, these results should be analyzed in detail and carefully.
In this study, 120 dairy farms with equal number of milk cooling tanks located in Northern Antioquia (Colombia) were sampled to determine the presence of ESBL-producing Enterobacteriaceae in bulk-tank milk and to explore the risk factors associated to its presence.
Table 5 Unconditional logistic regression analysis of risk factors associated with the presence of ESBL-producing Enterobacteriaceae in bulk-tank milk (n = 120) at Entrerríos, Antioquia (Colombia).
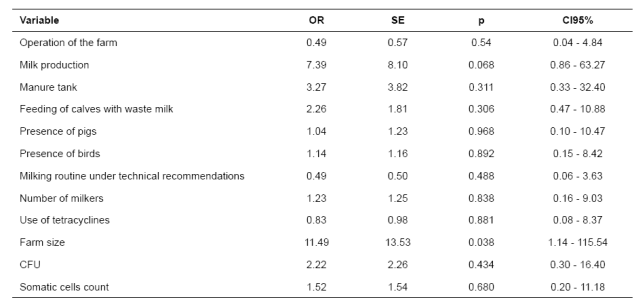
CFU: colony forming units; OR: odds ratio; SE: standard error; p value: significance level.
Recently, studies of the characteristics and prevalence of ESBL-producing Enterobacteriaceae in animals have increased in different countries worldwide (Smet et al., 2010; Ohnishi et al., 2013; Timofte et al., 2014; Skočková et al, 2015; Sudarwanto et al., 2015; Odenthal et al., 2016). However, relatively few studies have been conducted in Colombia on antibiotic resistance in food- producing animals and their products (Vanegas López et al., 2012; Jiménez Velásquez et al., 2013; Donado- Godoy et al., 2015) compared to other countries and to our knowledge, this is the first study conducted to determine the occurrence of ESBL-producing Enterobacteriaceae in bulk-tank milk in Colombia.
The frequency of ESBL-producing Enterobacteriaceae reported in this study (3.3%) agrees with other studies conducted in bulk-tank milk samples in Switzerland, Brazil, India, the Czech Republic, Indonesia, and Germany, which reported prevalences ranging between 0 and 9% (Geser et al., 2012; Nóbrega et al., 2013; Rasheed et al., 2014; Skočková et al., 2015; Sudarwanto et al., 2015; Odenthal et al., 2016).
Although the reported occurrences are low, they represent a significant finding that can contribute to clarifying the role that animals and their products have in the dissemination of multi-resistant bacteria in foods of animal origin.
The low frequency of ESBL-producing Enterobacteriaceae in bulk-tank milk could be a result of the high hygiene standards required by milk processing companies in Antioquia to purchase raw milk. Milk payment in Colombia is dependent on quality parameters such as CFU. Milk processing companies pay milk to producers as follows: CFU counts between 0 and 175,000 CFU/mL are considered as milk of very good quality and receive a bonus payment, CFU counts between 175,001 and 200,000 CFU/mL receive neither a bonus payment nor a discount on the price of milk, and counts
>200,001 CFU/mL receive discount on the price of milk (Ministerio de Agricultura, 2012). Improving the general hygiene in all stages of production and thereby reducing the microbial load of food products will also reduce the antimicrobial resistance load (Wegener, 2012). It is also possible that the low occurrence of ESBLs in these farms is due to little use of cephamycins and carbapenems since the selection pressure that drives ESBL evolution has usually been attributed to the intensive use of oxyimino-cephalosporins, mainly third-generation cephalosporins (Gniadkowski, 2001).
Our isolates contrast with the predominant isolates in dairy farms from other areas where E. coli and K. pneumoniae have been most frequently reported (Locatelli et al., 2010; Dahmen et al., 2013; Ohnishi et al., 2013; Sudarwanto et al., 2015; Odenthal et al., 2016). However, the relevance of other bacteria of the Enterobacteriaceae family could be underestimated because studies have mainly focused on E. coli and K. pneumoniae (Smet et al., 2010; Schmid et al., 2013; Sudarwanto et al., 2015; Odenthal et al., 2016). Anyway, the discovery of these bacteria (Enterobacter sp. and Serratia sp.) in milk is striking because usually Serratia fonticola is found in a wide array of environments, including drinking water, soil, and sewage (Aljorayid et al., 2016) while Enterobacter strains tend to colonize hospitalized patients, particularly those treated with antibiotics, and have been associated with infections of burns, wounds, respiratory and urinary tract (Puerta-García and Mateos-Rodríguez, 2010).
Although resistance to different antibiotics was detected, both by the disc diffusion method in agar (Kirby-Bauer) and by the microdilution broth method, sensitivity remained to carbapenem antibiotics. Carbapenems are the most potent beta- lactam antibiotics because of their wide spectrum (Bush, 2013), and these bacteria are still sensitive to this family of antibiotics, which makes a battery of effective antibiotics available for treating infections caused in humans and animals. However, carbapenems require responsible use to prevent the emergence of resistance.
The susceptibility patterns shown by both tests were similar. Discrepancies between results can be based on the pattern comparison strategy of the Vitek® automated expert system, which compares the observed phenotype with descriptions based on knowledge constructed from data obtained in scientific publications complemented by internal data of each health system (Gerst, 2000); if the system detects a discrepancy between that observed for an antibiotic and the distribution of MICs for the rest
of the antibiotics, it has the ability to modify the result, thus providing rapid, automatic and systematic validation of each of the sensitivity results while at the same time minimizing human error (Sanders et al., 2001; Schwaber et al., 2006; bioMérieux, 2015).
Given that production of most ESBLs is encoded by plasmids, co-resistance to other groups of antibiotics is common (Rybak et al., 2004; Xian- Zhi et al., 2007; Geser et al., 2012). The resistance shown to gentamicin and erythromycin by 100% of isolates possibly indicates the presence of a gene encoding resistance to antibiotics of aminoglycosides and macrolides families simultaneously. Notably, the range of antibiotics for which resistance was acquired is large and sufficiently worrisome because it shows the severity of the emergence of antibiotic resistance in different pathogens. For example, an infection caused by a strain of E. coli such as that found in our study could represent a larger problem because therapeutic options are clearly limited.
Recently, rapid growth in the number of positive isolates with CTX-M enzymes has been observed (Woodford, 2010). The CTX-M enzymes constitute a rapidly growing family of ESBLs enzymes with significant clinical impact (Zhao and Hu, 2013). Enterobacteriaceae isolates that are positive for CTX-M have been found in food-producing animals and their products (Carattoli, 2008; Smet et al., 2010; Schmid et al., 2013; Randall et al., 2014). The enzymes found in our isolates agree with those frequently reported worldwide. The CTX-M-96 (CTX-M-12a alternative name) is one of 109 variants of CTX-M enzymes that have been identified and assigned in the Lahey database (Zhao and Hu, 2013). The CTX-M-96 was reported in K. pneumoniae as host organism and contained in the GenBank® with the code AJ704396 (NCBI 2005). The CTX-M-96 was previously reported in K. pneumoniae in Chile and Argentina (NCBI, 2015; NCBI 2005). One study published this enzyme as a simulation model for evaluating activity towards oxyimino cephalosporins (Ghiglione et al., 2015). However, with its alternate name, the bla gene CTX-M-12a was detected first in Colombia in four isolates: two E. cloacae and in two K. pneumoniae isolates; one of the E. cloacae isolates and one of the K. pneumoniae isolates were associated with nosocomial infections while the other one was isolated from the community (Mantilla et al., 2009). A later study revealed that 18 out of 33 ESBL positive strains isolated from three Colombian hospitals harbored group 1 blaCTX-M and analysis of DNA sequences revealed the presence of blaCTX-M-12a in three of them (Ruiz et al., 2011). These results alert on the policies concerning antibiotic use, distribution, selling and management and other factors that may be associated with the spread of multiresistant organisms causing outbreaks in hospitals and the community, including the agricultural sector. The presence of the same gene at the hospital level and at the animal production level gives rise to conduct future studies to elucidate the relationship between both environments and behavior of antibiotic resistance in Colombia. It is important to conduct more molecular epidemiological studies of bacterial resistance that permit identifying genetic determinants and variants of the ESBL enzyme families that may be associated with changes in the spectrum and that decrease antimicrobial activity in a particular environment.
Relative to risk factors associated with the presence of ESBL-producing Enterobacteriaceae, the risk factors found in our study differ from those reported in previous studies in England and north of Wales, Switzerland and Israel (Snow et al., 2012; Reist et al., 2013; Adler et al., 2015) considering that independent variables are different between studies. In these studies, the following factors: use of third and fourth generation cephalosporins in the last 12 months, operating as an open farm, infrequent cleaning of equipment for feeding of calves and storage of fecal matter in manure tanks, animals originating from farms with primary production type dairy, animals originating from farms with more than one animal movement per day per 100, lack of a cooling system, increased crowdedness and lack of manure cleaning, antimicrobial prophylaxis and increased frequency of veterinarian visits, were associated with the presence of ESBL-producing Enterobacteriaceae in cattle (Snow et al., 2012; Reist et al., 2013; Adler et al., 2015).
The risk factor dairy farm size associated with the presence of ESBL-producing Enterobacteriaceae in our study could be explained by the fact that a larger number of milking cows could represent an increase in the circulation and occurrence of diseases in a
farm, which would finally lead to the frequent use of antibiotics (Kreausukon, 2011) and/or increased contact between milkers and cows that are sick or being treated. Similarly, the larger number of cows in milking could lead to non-compliance of the stringent routine milking standards between cows (Ruiz-Romero, 2015)
The fact that most of the farms had reported performing milking routine following technical recommendations favors the sanitary situation in the municipality because if there is no monitoring of good milking practices, there would be more cases of mastitis and therefore a greater usage of antibiotics with the possibility of developing or acquiring resistance (Oliver and Murinda, 2012). In the same way, situations in which the choice and provision of antibiotics is in the hands of a person other than the veterinarian may cause variations in bacterial sensitivity along with recurrence of the presentation of microorganisms associated with different pathologies as a consequence of not normally performing isolation or evaluation by antibiogram of the causal agent of infection (Betancourt et al., 2003).
In this study, feeding calves with waste milk (milk of cows with antibiotic treatment) -although not found as a risk factor- continues to be a frequent activity in dairy farms of the municipality, despite current knowledge of the association between this practice and increased resistance to antimicrobials in calves (Langford et al., 2003; Aust et al., 2013; Brunton et al., 2014).
The use of bulk-tank milk samples could serve to monitor trends in resistance to antibiotics in dairy farms (Berge et al., 2007). Although the presence of ESBL-producing Enterobacteriaceae was not high in this study, the use of antibiotics should be controlled, sanitary measures should be reinforced, and the phenomenon of antibiotic resistance in food- producing animals should continue to be investigated (FDA, 2012). Timely identification of ESBL- producing Enterobacteriaceae strains is essential in the monitoring of the development of antimicrobial resistance and in the implementation of infection control measures for the protection of public health (WHO, 2001).
Acknowledgments
This study was funded by the Fondo de Apoyo al Primer Proyecto de Profesores of the Universidad de Antioquia (Comité para el Desarrollo de la Investigación -CODI). Authors thank the Laboratorio de Microbiología Veterinaria of the Facultad de Ciencias Agrarias (UdeA), María Luisa Vélez Agudelo (joven investigadora CODI, 2013), the Estrategia de Sostenibilidad 2013-2014, CENTAURO, the Secretaría de Asistencia Técnica Agropecuaria y Desarrollo Comunitario de Entrerríos, and participating dairy herds.